DOI: 10.31038/JMG.2020321
Short Communication
Recently, a very powerful web-server predictor has been established for identifying the subcellular localization of a protein based on its sequence information alone for the multi-label systems 1], in which a same protein may occur or move between two or more location sites and hence needs to be marked with the multi-label approach 2]. The web-server predictor is called “pLoc_bal-mEuk”, where “bal” means the web-server has been further improved by the “balance treatment” 3-9], and “m” means the capacity able to deal with the multi-label systems. To find how the web-server is working, please do the following.
Click the link at http://www.jci-bioinfo.cn/pLoc_bal-mEuk/, the top page of the pLoc_bal-mEuk web-server will appear on your computer screen, as shown in Figure 1. Then by following the Step 2 and Step 3 in 5], you will see the predicted results shown on Figure 2. Nearly all the success rates achieved by the web-server predictor for the eukaryotic proteins in each of the 22 subcellular locations are within the range of 90-100%, which is far beyond the reach of any of its counterparts.
Figure 1. A semi screenshot for the top page of pLoc_bal-mEuk (Adapted from 5]).
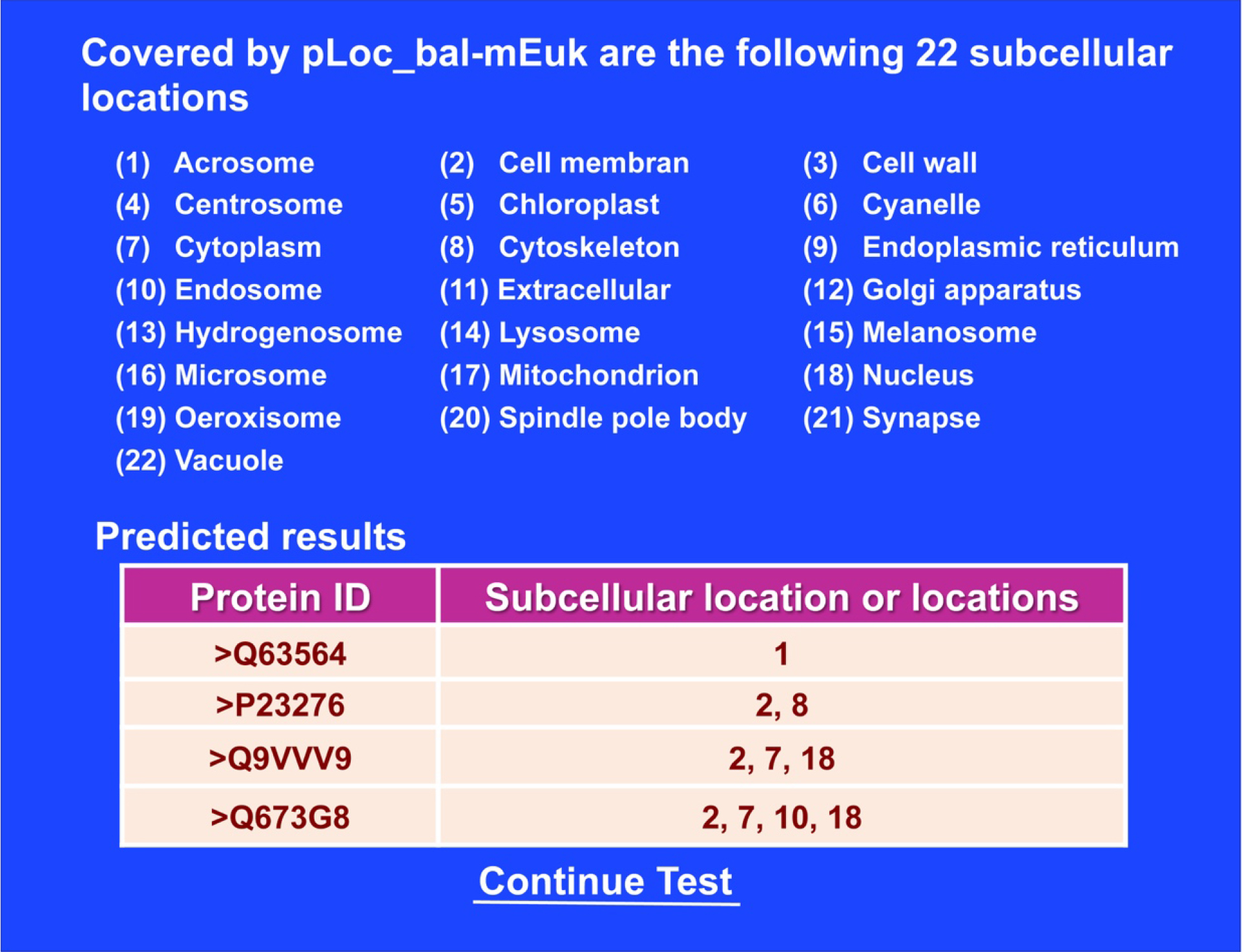
Figure 2. A semi screenshot for the webpage obtained by following Step 3 of Section 3.5 (Adapted from 5]).
Besides, the web-server predictor has been developed by strictly observing the guidelines of “Chou’s 5-steps rule” and hence have the following notable merits (see, e.g., 10-90] and three comprehensive review papers 2, 91, 92]: (1) crystal clear in logic development, (2) completely transparent in operation, (3) easily to repeat the reported results by other investigators, (4) with high potential in stimulating other sequence-analyzing methods, and (5) very convenient to be used by the majority of experimental scientists.
For the fantastic and awesome roles of the “5-steps rule” in driving proteome, genome analyses and drug development, see a series of recent papers [2, 92-103] where the rule and its wide applications have been very impressively presented from various aspects or at different angles.
References
- Chou KC, Shen HB (2007) Recent progresses in protein subcellular location prediction. Analytical Biochemistry 370: 1–16. [Crossref]
- Chou KC (2019) Advance in predicting subcellular localization of multi-label proteins and its implication for developing multi-target drugs. Current Medicinal Chemistry 6: 4918–4943. [Crossref]
- Xiao X, Cheng X, Chen G, Mao Q, Chou KC (2019) pLoc_bal-mVirus: Predict Subcellular Localization of Multi-Label Virus Proteins by Chou’s General PseAAC and IHTS Treatment to Balance Training Dataset. Med Chem 15: 496–509.
- Xiao X, Cheng X, Chen G, Mao Q, Chou KC (2019) pLoc_bal-mGpos: predict subcellular localization of Gram-positive bacterial proteins by quasi-balancing training dataset and PseAAC. Genomics 111: 886–892. [Crossref]
- Chou KC, Cheng X, Xiao X (2019) pLoc_bal-mEuk: predict subcellular localization of eukaryotic proteins by general PseAAC and quasi-balancing training dataset. Med Chem 15: 472–485. [Crossref]
- Chou KC, Cheng X, Xiao X (2019) pLoc_bal-mHum: predict subcellular localization of human proteins by PseAAC and quasi-balancing training dataset. Genomics 111: 1274–1282. [Crossref]
- Cheng X, Lin WZ, Xiao X, Chou KC (2019) pLoc_bal-mAnimal: predict subcellular localization of animal proteins by balancing training dataset and PseAAC. Bioinformatics 35: 398–406. [Crossref]
- Cheng X, Xiao X, Chou KC (2018) pLoc_bal-mPlant: predict subcellular localization of plant proteins by general PseAAC and balancing training dataset. Curr Pharm Des 24: 4013–4022. [Crossref]
- Cheng X, Xiao X, Chou KC (2018) pLoc_bal-mGneg: predict subcellular localization of Gram-negative bacterial proteins by quasi-balancing training dataset and general PseAAC. Journal of Theoretical Biology 458: 92–102. [Crossref]
- Butt AH, Khan YD (2018) Prediction of S-Sulfenylation Sites Using Statistical Moments Based Features via Chou’s 5-Step Rule. International Journal of Peptide Research and Therapeutics (IJPRT).
- Awais M, Hussain W, Khan YD, Rasool N, Khan SA, et al. (2019) iPhosH-PseAAC: Identify phosphohistidine sites in proteins by blending statistical moments and position relative features according to the Chou’s 5-step rule and general pseudo amino acid composition. IEEE/ACM Trans Comput Biol Bioinform 27. [Crossref]
- Barukab O, Khan YD, Khan SA, Chou KC (2019) iSulfoTyr-PseAAC: Identify tyrosine sulfation sites by incorporating statistical moments via Chou’s 5-steps rule and pseudo components. Current Genomics 20.
- AH Butt, YD Khan (2019) Prediction of S-Sulfenylation Sites Using Statistical Moments Based Features via Chou’s 5-Step Rule. International Journal of Peptide Research and Therapeutics (IJPRT)
- Chen Y, Fan X (2019) Use of Chou’s 5-Steps Rule to Reveal Active Compound and Mechanism of Shuangshen Pingfei San on Idiopathic Pulmonary Fibrosis. Current Molecular Medicine 11 [Crossref]
- Du X, Diao Y, Liu H, Li S (2019) MsDBP: Exploring DNA-binding Proteins by Integrating Multi-scale Sequence Information via Chou’s 5-steps Rule. Journal of Proteome Research 18: 3119–3132. [Crossref]
- Ehsan A, Mahmood MK, Khan YD, Barukab OM, Khan SA, et al. (2019) iHyd-PseAAC (EPSV): Identify hydroxylation sites in proteins by extracting enhanced position and sequence variant feature via Chou’s 5-step rule and general pseudo amino acid composition. Current Genomics 20: 124–133. [Crossref]
- Hussain W, Khan SD, Rasool N, Khan SA, Chou KC (2019) SPalmitoylC-PseAAC: A sequence-based model developed via Chou’s 5-steps rule and general PseAAC for identifying S-palmitoylation sites in proteins. Anal Biochem 568: 14–23. [Crossref]
- Hussain W, Khan YD, Rasool N, Khan SA, Chou KC (2019) SPrenylC-PseAAC: A sequence-based model developed via Chou’s 5-steps rule and general PseAAC for identifying S-prenylation sites in proteins. J Theor Biol 468: 1–11. [Crossref]
- Ju Z, Wang SY (2020) Prediction of lysine formylation sites using the composition of k-spaced amino acid pairs via Chou’s 5-steps rule and general pseudo components. Genomics 112: 859–866. [Crossref]
- Kabir M, Ahmad S, Iqbal M, Hayat M (2020) iNR-2L:A two-level sequence-based predictor developed via Chou’s 5-steps rule and general PseAAC for identifying nuclear receptors and their families. Genomics 112: 276–285. [Crossref]
- Khan ZU, Ali F, Khan IA, Hussain Y, Pi D (2019) iRSpot-SPI: Deep learning-based recombination spots prediction byincorporating secondary sequence information coupled withphysio-chemical properties via Chou’s 5-step rule and pseudo components. Chemometrics and Intelligent Laboratory Systems (CHEMOLAB) 189: 169–180.
- Lan J, Liu J, Liao C, Merkler DJ, Han Q, et al. (2019) A Study for Therapeutic Treatment against Parkinson’s Disease via Chou’s 5-steps Rule. Current Topics in Medicinal Chemistry 19.
- Le NQK (2019) iN6-methylat (5-step): identifying DNA N(6)-methyladenine sites in rice genome using continuous bag of nucleobases via Chou’s 5-step rule. Mol Genet Genomics 294: 1173–1182. [Crossref]
- Le NQK, Yapp EKY, Ho QT, Nagasundaram N, Ou YY, et al. (2019) iEnhancer-5Step: Identifying enhancers using hidden information of DNA sequences via Chou’s 5-step rule and word embedding. Anal Biochem 571: 53–61. [Crossref]
- Le NQK, Yapp EKY, Ou YY, Yeh HY (2019) iMotor-CNN: Identifying molecular functions of cytoskeleton motor proteins using 2D convolutional neural network via Chou’s 5-step rule. Anal Biochem 575: 17–26. [Crossref]
- Liang R, Xie J, Zhang C, Zhang M, Huang H, et al. (2019) Identifying Cancer Targets Based on Machine Learning Methods via Chou’s 5-steps Rule and General Pseudo Components. Current Topics in Medicnal Chemistry 19: 2301–2317. [Crossref]
- Liang Y, Zhang S, Identifying DNase I hypersensitive sites using multi-features fusion and F-score features selection via Chou’s 5-steps rule. Biophys Chem 253: 106227. [Crossref]
- Malebary SJ, Rehman MSU, Khan YD (2019) iCrotoK-PseAAC: Identify lysine crotonylation sites by blending position relative statistical features according to the Chou’s 5-step rule. PLoS One 14: 0223993.
- Nazari I, Tahir M, Tayari H, Chong KT (2019) iN6-Methyl (5-step): Identifying RNA N6-methyladenosine sites using deep learning mode via Chou’s 5-step rules and Chou’s general PseKNC. Chemometrics and Intelligent Laboratory Systems (CHEMOLAB) 193: 103811.
- Ning Q, Ma Z, Zhao X (2019) dForml(KNN)-PseAAC: Detecting formylation sites from protein sequences using K-nearest neighbor algorithm via Chou’s 5-step rule and pseudo components. J Theor Biol 470: 43–49. [Crossref]
- Salman, Khan M, Iqbal N, Hussain T, Afzal S, et al. (2019) A two-level computation model based on deep learning algorithm for identification of piRNA and their functions via Chou’s 5-steps rule. International Journal of Peptide Research and Therapeutics (IJPRT).
- Tahir M, Tayara H, Chong KT (2019) iDNA6mA (5-step rule): Identification of DNA N6-methyladenine sites in the rice genome by intelligent computational model via Chou’s 5-step rule. CHEMOLAB 189: 96–101.
- Vishnoi S, Garg P, Arora P (2020) Physicochemical n-Grams Tool: A tool for protein physicochemical descriptor generation via Chou’s 5-steps rule. Chem Biol Drug Des 95: 79–86. [Crossref]
- Wiktorowicz A, Wit A, Dziewierz A, Rzeszutko L, Dudek D, et al. (2019) Calcium Pattern Assessment in Patients with Severe Aortic Stenosis Via the Chou’s 5-Steps Rule. Current Pharmaceutical Design 25: 3769–3775. [Crossref]
- Yang L, Lv Y, Wang S, Zhang Q, Pan Y, et al. (2019) Identifying FL11 subtype by characterizing tumor immune microenvironment in prostate adenocarcinoma via Chou’s 5-steps rule. Genomics [Crossref]
- Khan YD, Amin N, Hussain W, Rasool N, Khan SA, et al. (2020) iProtease-PseAAC(2L): A two-layer predictor for identifying proteases and their types using Chou’s 5-step-rule and general PseAAC. Anal Biochem 588: 113477.
- Xu Y, Ding J, Wu LY, Chou KC (2013) iSNO-PseAAC: Predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition PLoS ONE 8: 55844. [Crossref]
- Xu Y, Shao XJ, Wu LY, Deng NY, Chou KC (2013) iSNO-AAPair: incorporating amino acid pairwise coupling into PseAAC for predicting cysteine S-nitrosylation sites in proteins. PeerJ 1: 171. [Crossref]
- Xu Y, Wen X, Shao XJ, Deng NY, Chou KC (2014) iHyd-PseAAC: Predicting hydroxyproline and hydroxylysine in proteins by incorporating dipeptide position-specific propensity into pseudo amino acid composition. International Journal of Molecular Sciences (IJMS) 15: 7594–7610. [Crossref]
- Yan Xu, Xin Wen, Li-Shu Wen, Ling-Yun Wu, Nai-Yang Deng, et al. (2014) iNitro-Tyr: Prediction of nitrotyrosine sites in proteins with general pseudo amino acid composition. PLoS ONE 9: 105018.
- Xu Y, Chou KC (2016) Recent progress in predicting posttranslational modification sites in proteins. Curr Top Med Chem 16: 591–603. [Crossref]
- Liu LM, Xu Y, Chou KC (2017) iPGK-PseAAC: identify lysine phosphoglycerylation sites in proteins by incorporating four different tiers of amino acid pairwise coupling information into the general PseAAC. Med Chem 13: 552–559. [Crossref]
- Xu Y, Wang Z, Li C, Chou KC (2017) iPreny-PseAAC: identify C-terminal cysteine prenylation sites in proteins by incorporating two tiers of sequence couplings into PseAAC. Med Chem 13: 544–551. [Crossref]
- Cai L, Wan CL, He L, Jong De S, Chou KC (2015) Gestational influenza increases the risk of psychosis in adults. Medicinal Chemistry 11: 676–682. [Crossref]
- Liu J, Song J, Wang MY, He L, Cai L et al. (2015) Association of EGF rs4444903 and XPD rs13181 polymorphisms with cutaneous melanoma in Caucasians. Medicinal Chemistry 11: 551–559. [Crossref]
- Cai L, Yang YH, He L, Chou KC (2016) Modulation of cytokine network in the comorbidity of schizophrenia and tuberculosis. Curr Top Med Chem 16: 655–665. [Crossref]
- Lei Cai, Wei Yuan, Zhou Zhang, Lin He, Kuo-Chen Chou (2016) In-depth comparison of somatic point mutation callers based on different tumor next-generation sequencing depth data. Scientific Reports 6: 36540.
- Zhu Y, Cong QW, Liu Y, Wan CL, Yu T, et al. (2016) Antithrombin is an importantly inhibitory role against blood clots. Curr Top Med Chem 16: 666–674.
- Zhang ZD, Liang K, Li K, Wang GQ, Zhang KW, et al. (2017) Chlorella vulgaris induces apoptosis of human non-small cell lung carcinoma (NSCLC) cells. Med Chem 13: 560–568. [Crossref]
- Cai L, Huang T, Su J, Zhang X, Chen W, et al. (2018) Implications of newly identified brain eQTL genes and their interactors in Schizophrenia. Molecular Therapy – Nucleic Acids 12: 433–442. [Crossref]
- Niu B, Zhang M, Du P, Jiang L, Qin R, et al. (2017) Small molecular floribundiquinone B derived from medicinal plants inhibits acetylcholinesterase activity. Oncotarget 8: 57149–57162. [Crossref]
- Su Q, Lu W, Du D, Chen F, Niu B, et al. (2017) Prediction of the aquatic toxicity of aromatic compounds to tetrahymena pyriformis through support vector regression. Oncotarget 8: 49359–49369. [Crossref]
- Yi Lu, Shuo Wang, Jianying Wang, Guangya Zhou, Qiang Zhang, et al. (2019) An Epidemic Avian Influenza Prediction Model Based on Google Trends. Letters in Organic Chemistry 16: 303–310.
- Niu B, Liang C, Lu Y, Zhao M, Chen Q, et al. (2019) Glioma stages prediction based on machine learning algorithm combined with protein-protein interaction networks. Genomics 112: 837–847. [Crossref]
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) Identification of protein-protein binding sites by incorporating the physicochemical properties and stationary wavelet transforms into pseudo amino acid composition (iPPBS-PseAAC). J Biomol Struct Dyn (JBSD) 34: 1946–1961. [Crossref]
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) iSuc-PseOpt: Identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal Biochem 497: 48–56. [Crossref]
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) pSuc-Lys: Predict lysine succinylation sites in proteins with PseAAC and ensemble random forest approach. Journal of Theoretical Biology 394: 223–230. [Crossref]
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) iCar-PseCp: identify carbonylation sites in proteins by Monto Carlo sampling and incorporating sequence coupled effects into general PseAAC. Oncotarget 7: 34558–34570. [Crossref]
- Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) iPPBS-Opt: A Sequence-Based Ensemble Classifier for Identifying Protein-Protein Binding Sites by Optimizing Imbalanced Training Datasets. Molecules 21: 95. [Crossref]
- Jia J, Zhang L, Liu Z, Xiao X, Chou KC (2016) pSumo-CD: Predicting sumoylation sites in proteins with covariance discriminant algorithm by incorporating sequence-coupled effects into general PseAAC. Bioinformatics 32: 3133–3141. [Crossref]
- Liu Z, Xiao X, Yu DJ, Jia J, Qiu WR, et al. (2016) pRNAm-PC: Predicting N-methyladenosine sites in RNA sequences via physical-chemical properties. Anal Biochem 497: 60–67. [Crossref]
- Xiao X, Ye HX, Liu Z, Jia JH, Chou KC (2016) iROS-gPseKNC: predicting replication origin sites in DNA by incorporating dinucleotide position-specific propensity into general pseudo nucleotide composition. Oncotarget 7: 34180–34189. [Crossref]
- Qiu WR, Sun BQ, Xiao X, Xu ZC, Jia JH, et al. (2018) iKcr-PseEns: Identify lysine crotonylation sites in histone proteins with pseudo components and ensemble classifier. Genomics 110: 239–246. [Crossref]
- Jia J, Li X, Qiu W, Xiao X, Chou KC (2019) iPPI-PseAAC(CGR): Identify protein-protein interactions by incorporating chaos game representation into PseAAC. Journal of Theoretical Biology 460: 195–203.
- Chen W, Ding H, Feng P, Lin H, Chou KC (2016) iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget 7: 16895–16909. [Crossref]
- Chen W, Feng P, Ding H, Lin H, Chou KC (2016) Using deformation energy to analyze nucleosome positioning in genomes. Genomics 107: 69–75. [Crossref]
- Chen W, Tang H, Ye J, Lin H, Chou KC (2016) iRNA-PseU: Identifying RNA pseudouridine sites. Molecular Therapy – Nucleic Acids 5: 332. [Crossref]
- Chang-Jian Zhang, Hua Tang, Wen-Chao Li, Hao Lin, Wei Chen, et al. (2016) iOri-Human: identify human origin of replication by incorporating dinucleotide physicochemical properties into pseudo nucleotide composition. Oncotarget 7: 69783–69793.
- Chen W, Feng P, Yang H, Ding H, Lin H, et al. (2017) iRNA-AI: identifying the adenosine to inosine editing sites in RNA sequences. Oncotarget 8: 4208–4217. [Crossref]
- Feng P, Ding H, Yang H, Chen W, Lin H, et al. (2017) iRNA-PseColl: Identifying the occurrence sites of different RNA modifications by incorporating collective effects of nucleotides into PseKNC. Molecular Therapy – Nucleic Acids 7: 155–163. [Crossref]
- Chen W, Ding H, Zhou X, Lin H, Chou KC (2018) iRNA(m6A)-PseDNC: Identifying N6-methyladenosine sites using pseudo dinucleotide composition. Analytical Biochemistry 561–562: 59–65. [Crossref]
- Chen W, Feng P, Yang H, Ding H, Lin H, et al. (2018) iRNA-3typeA: identifying 3-types of modification at RNA’s adenosine sites. Molecular Therapy: Nucleic Acid 11: 468–474. [Crossref]
- Su ZD, Huang Y, Zhang ZY, Zhao YW, Wang D, et al. (2018) iLoc-lncRNA: predict the subcellular location of lncRNAs by incorporating octamer composition into general PseKNC. Bioinformatics 34: 4196–4204. [Crossref]
- Yang H, Qiu WR, Liu G, Guo FB, Chen W, et al. (2018) iRSpot-Pse6NC: Identifying recombination spots in Saccharomyces cerevisiae by incorporating hexamer composition into general PseKNC. International Journal of Biological Sciences 14: 883–891. [Crossref]
- Feng P, Yang H, Ding H, Lin H, Chen W, et al. (2019) iDNA6mA-PseKNC: Identifying DNA N(6)-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics 111: 96–102. [Crossref]
- Du QS, Wang SQ, Xie NZ, Wang QY, Huang RB, et al. (2017) 2L-PCA: A two-level principal component analyzer for quantitative drug design and its applications. Oncotarget 8: 70564–70578. [Crossref]
- Liu B, Fang L, Long R, Lan X, Chou KC (2016) iEnhancer-2L: a two-layer predictor for identifying enhancers and their strength by pseudo k-tuple nucleotide composition. Bioinformatics 32: 362–369. [Crossref]
- Liu B, Long R, Chou KC (2016) iDHS-EL: Identifying DNase I hypersensi-tivesites by fusing three different modes of pseudo nucleotide composition into an ensemble learning framework. Bioinformatics 32: 2411–2418. [Crossref]
- Liu B, Wang S, Long R, Chou KC (2017) iRSpot-EL: identify recombination spots with an ensemble learning approach. Bioinformatics 33: 35–41. [Crossref]
- Liu B, Yang F, Chou KC (2017) 2L-piRNA: A two-layer ensemble classifier for identifying piwi-interacting RNAs and their function. Molecular Therapy – Nucleic Acids 7: 267–277.
- Liu B, Li K, Huang DS, Chou KC (2018) iEnhancer-EL: Identifying enhancers and their strength with ensemble learning approach. Bioinformatics 34: 3835–3842. [Crossref]
- Liu B, Weng F, Huang DS, Chou KC (2018) iRO-3wPseKNC: Identify DNA replication origins by three-window-based PseKNC. Bioinformatics 34: 3086–3093. [Crossref]
- Liu B, Yang F, Huang DS, Chou KC (2018) iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics 34: 33–40. [Crossref]
- Qiu WR, Sun BQ, Xiao X, Xu ZC, Chou KC (2016) iHyd-PseCp: Identify hydroxyproline and hydroxylysine in proteins by incorporating sequence-coupled effects into general PseAAC. Oncotarget 7: 44310–44321. [Crossref]
- Qiu WR, Sun BQ, Xiao X, Xu ZC, Chou KC (2016) iPTM-mLys: identifying multiple lysine PTM sites and their different types. Bioinformatics 32: 3116–3123. [Crossref]
- Qiu WR, Xiao X, Xu ZC, Chou KC (2016) iPhos-PseEn: identifying phosphorylation sites in proteins by fusing different pseudo components into an ensemble classifier. Oncotarget 7: 51270–51283. [Crossref]
- Qiu WR, Jiang SY, Sun BQ, Xiao X, Cheng X, et al. (2017) iRNA-2methyl: identify RNA 2′-O-methylation sites by incorporating sequence-coupled effects into general PseKNC and ensemble classifier. Medicinal Chemistry 13: 734–743. [Crossref]
- Qiu WR, Jiang SY, Xu ZC, Xiao X, Chou KC (2017) iRNAm5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget 8: 41178–41188. [Crossref]
- Qiu WR, Sun BQ, Xiao X, Xu D, Chou KC (2017) iPhos-PseEvo: Identifying human phosphorylated proteins by incorporating evolutionary information into general PseAAC via grey system theory. Molecular Informatics 36. [Crossref]
- Zhai X, Chen M, Lu W (2018) Accelerated search for perovskite materials with higher Curie temperature based on the machine learning methods. Computational Materials Science 151: 41–48.
- Chou KC (2011) Some remarks on protein attribute prediction and pseudo amino acid composition (50th Anniversary Year Review, 5-steps rule). Journal of Theoretical Biology 273 236–247. [Crossref]
- Chou KC (2019) Impacts of pseudo amino acid components and 5-steps rule to proteomics and proteome analysis. Current Topics in Medicinal Chemistry (CTMC) 19.
- Chou KC (2019) Two kinds of metrics for computational biology. Genomics.
- Chou KC (2019) Proposing pseudo amino acid components is an important milestone for proteome and genome analyses. International Journal for Peptide Research and Therapeutics (IJPRT).
- Chou KC (2019) An insightful recollection for predicting protein subcellular locations in multi-label systems. Genomics.
- Chou KC (2019) Progresses in predicting post-translational modification. International Journal of Peptide Research and Therapeutics (IJPRT).
- Chou KC (2019) Recent Progresses in Predicting Protein Subcellular Localization with Artificial Intelligence (AI) Tools Developed Via the 5-Steps Rule. Japanese Journal of Gastroenterology and Hepatology 2: 1–4.
- Chou KC (2019) An insightful recollection since the distorted key theory was born about 23 years ago. Genomics.
- Chou KC (2019) Artificial intelligence (AI) tools constructed via the 5-steps rule for predicting post-translational modifications. Trends in Artificial Inttelengence (TIA) 3: 60–74.
- Chou KC (2020) Distorted Key Theory and Its Implication for Drug Development. Current Genomics.
- Chou KC (2019) An Insightful 10-year Recollection Since the Emergence of the 5-steps Rule. Current Pharmaceutical Design 4223–4234.
- Chou KC (2019) An insightful recollection since the birth of Gordon Life Science Institute about 17 years ago. Advancement in Scientific and Engineering Research 4: 31–36.
- Chou KC (2019) Gordon Life Science Institute: Its philosophy, achievements, and perspective. Annals of Cancer Therapy and Pharmacology 2: 001–26.