Abstract
The current ongoing coronavirus disease 2019 (COVID-19 SARS2 and detection and isolation of infected people with the virus is crucial. Real-time polymerase chain reaction (RtPCR) test has proven to be most useful in viral detection. The objective of this work is to determine the benefit of group testing for resource optimization and be able to expand the number of individuals who are screened to find healthy COVID-19 carriers. Groups from 2 to 27 of a total of 2,100 people were included. RT-PCR was done with Seegene kits for mRNA extraction and reagents in Biorad RT-PCR machine. Results: groups of between 4 to 6 people have been the ones with the best optimization results with 64 to 67%, this means that with 1 test it is possible to cover the same number of people and detect more quickly those who must be isolated and follow their contacts. Conclusion: Grouping tests can optimize resources up to 67%.
Background
The current ongoing coronavirus disease 2019 (COVID-19 SARS2) pandemic with mainly severe acute respiratory syndrome, is a serious global public health problem. The detection and isolation of infected people with the virus is crucial. Hitherto, the real-time polymerase chain reaction (RT-PCR) test has proven to be most useful in viral detection when performed using nasal and oropharyngeal exudate samples. Due to the global and rapidly progressive nature of the pandemic, the tests have been limited in supply with a high cost; therefore, their optimization is important. The Many millions of tests have been performed almost exclusively on an individual basis and are many millions of tests [1]. To date, the United Kingdom has performed the highest number of tests globally, with 486 billion tested inhabitants [2]. In the open population, the first positivity reported by South Korea, who had performed the greatest number of tests, the statistic of positives was 1.7% in the open population, and later in May 2020 they performed 10 million tests and only had 300 positives (positivity rate of 0.9% of positivity) between 1,553,5523.. In the United States of America, the positivity rate in the suspected sample population was 19.8% and 9.6% in the general population of 32,009,840 by July 25, 20204. Based on this percentage, the negativity rate in the general population is expected to be high. It is known that a person infected by COVID-19 (SARS- CoV-2) who is asymptomatic, has an approximate viral load of 6.76 x 106 viral copies in the first five days of infection [3-9]. Subsequently, the viral load decreases from day 6, to 3.44 x 10 viral copies. In addition, it has been described that the viral load is not detectable by day 28 in up to 39.93% of infected people. On the other hand, patients with symptoms can present a viral load of up to 7.11 x 108 viral copies [5-9]. Taking these findings into account, it was found feasible to do group tests (pooling), the first published antecedent for COVID-19 is from Yelin and collaborators [10] in which they make groups of 35 people. In later publications, theoretical calculations are made on the number of people to include in the groups and with automated equipment [11-13], finding savings and optimization of resources of 69% [13].
Objective
Determine the benefit of group testing for resource optimization and be able to expand the number of individuals who are screened to find healthy COVID-19 carriers. In our laboratory to assess this observation at the beginning, we carried out a preliminary test with 19 people including technicians, doctors, administrative and cleaning staff, all asymptomatic, a sample was taken from each person and the pilot group was carried out, gathering aliquot of 19 people in a pool, performing a single RT-RCR test for COVID-19, obtaining a negative result. None developed any symptoms of COVID-19 in the next 2 months. Additionally, the same procedure was performed adding to that pool with an already known positive sample, obtaining the positive result in the pool sample.
Material and Methods
An observational, cross-sectional and descriptive study was carried out.
Selection and Inclusion Criteria
Open population with asymptomatic individuals who are considered healthy, including asymptomatic carriers, which is the objective of this study. Selection of people and groups: a call was made to individuals and companies of various types, who were named as suppliers: 1) Family group: people who live in the same home; 2) Work group: people who work in the same place; 3) Health group: health workers with or without direct contact with patients. The selection of the groups was initially proposed in groups selected by each supplier, without exceeding 30 people. People were recruited between May 21 and August 5, 2020. Exclusion criteria: People with respiratory symptoms of any type and degree and with fever were ruled out. Taking samples: It was carried out either in the laboratory or at the work sites. In both cases with the safety measures by the sampling personnel according to the WHO recommended. The transport was carried out with triple packaging. A nasopharyngeal sample was taken from each individual and placed in a tube with 2.0 ml of transport medium, from which an aliquot of 0.10 ml was taken from each individual to form the pool of that group, a single mRNA extraction was performed in each group. The samples were kept refrigerated for the first 24 hours and after that time they were kept frozen at -20°C until processing.
Sample Processing
Extraction of viral DNA/RNA, using Invisorb® Spin universal kit reagents Invitek Molecular Gmbh Berlin. In a 2 ml Safe lock tube, place 200 µl of the sample, with 200 µl HTL buffer, 20 µl of carrier DNA and 20 µl of Proteinase K, mix in vortex for 10 seconds, place the tube in thermomix and incubate with constant shaking x 10 minutes at 65°C and then 10 minutes at 95°C. Binding Add 260 µl of binding solution to the sample lysate and mix by pipetting up and down or vortex. The sample is incubated for 5 minutes at room temperature. The sample is transferred to the RTA Spin filter with the receiving RTA tube. The tube is closed and centrifuged for 1 minute at 11.1100 g. Discard the RTA Receiver with the filtrate and fit a new receiver tube. The samples are centrifuged at -20°C. Separately, the following reagents are prepared: 1) 5 µl of 2019-nCoV MOM reagent (Seegene®); 2) 5 µl of RNase-free water; 3) 5 μl of Buffer 5X Real-time One-Step (Seegene®), and 4) 2 μl of Real-time One-Step enzyme (Seegene®). The mixture is centrifuged briefly and 17 μl of One-step RT-PCR Mastermix (Seegene®) will be added.
8 μl of the group of samples, 2019-nCoV PC and RNAs-free distilled water will be added to the previous solution. Afterwards, the samples will be centrifuged and they will be placed in the equipment for RT-PCR. (2) The AllplexTM 2019-nCoV kit (Cat. No. RP10243X; Seegene®) will be used for the qualitative detection of SARS-CoV-2 (COVID-19). The kit evaluates the E, RdRP and N genes of SARS- CoV-2. (3) Polymerase chain reaction with reverse transcriptase. The samples will be placed in the equipment for RT-PCR (CFX96TM, BioRad®), with the following parameters: Cycle 1) Temperature 50°C, duration 20 minutes; Cycle 2) Temperature 95°C, duration 15 minutes; Cycles 3-44) Temperature 94°C, duration 15 seconds each; Cycle 45) Temperature 58°C, duration 30 seconds. The cycling determined to evaluate the positivity of the sample is 45 cycles according to the manufacturer’s protocol.
Data Interpretation
The data obtained from the RT-PCR process will be automatically analyzed in the CFX96 ManagerTM program included in the BioRad® kit. The cut-off point to establish a sample as positive is the value of Ct ≤40.
Statistical Analysis
Frequencies and percentages were determined, Pearson’s correlation test was applied. The data analysis was carried out with the SPSSv23 program. A p value <0.05 was considered as statistical significance. Ethical and biosafety aspects: The protocol was approved by the research department of the General Hospital of Mexico Eduardo Liceaga of CDMX with number DI / 20/405103150.
Results
2,100 participants were included, 60% (1,260 people) were men and 40% (840 people) were women. The average age was 35 years, with a range of 18 to 65 years. The participants were gathered into 536 groups, which meant performing the same number of tests. The groups consisted of a minimum of 2 people and a maximum of 23 people and were selected by the suppliers (Table 1), 17.35% (93 groups) were positive with a total of 131 positive people, which means 6.2% of positive people out of the 2,100 included in the study. To detect positive people in each group, 372 additional tests were required, optimization of the test resource in total groups between 2 to 27 people was 56.76%, this figure was obtained in relation to the total number of people included that would be equivalent to the tests performed individually (n 2,100, 100%) and the number of tests actually performed, obtained from the sum of the number of groups (n 536) plus the number of additional tests, of each positive group individual tests were performed in groups. The additional tests where done in the positive groups as follows: from 2 to 7 people individually and in those from 8 to 27 in subgroups of 2 or 3 and of them the positive ones individually, obtaining a total of 372 additional tests, which added to the initial ones were 908, the % optimization in relation to the Initial theoretical of 2,100 tests and the performed test 908 equivalent to 56.76% (100-(908 * 100/2100). Table 1 shows the % optimization for each group, the small % was 2 people and the better between 4, 5 and 6 people. There were between 1 and 2 groups of 8, 9 and 11 to 23 people who were carried out very early in the study (May- June) that had a very high % optimization (87 to 94) and it should be considered that at the beginning of the pandemic there were very few infected people. The number of members of each group had a low positive correlation with the number of additional tests performed (rho=0.13, p=0.009), while the number of additional tests performed had a high positive correlation with the number of tests that resulted positive for COVID-19 (rho=0.99, p=0.0001) (Figure 1) [14].
Table 1: Optimization% for each group.
|
People per group |
Total no. of people in the groups | n of groups | Positive no. of groups | no. of positive People | Additional tests | Total of Tests performed | Theoretically individual tests | % Optimization |
| 2 | 482 | 241 | 48 | 49 | 96 | 337 | 482 |
30.08 |
|
3 |
132 | 44 | 8 | 8 | 24 | 68 | 132 | 48.48 |
| 4 | 220 | 55 | 6 | 11 | 24 | 79 | 220 |
64.09 |
|
5 |
580 | 116 | 15 | 32 | 75 | 191 | 580 | 67.07 |
| 6 | 294 | 49 | 8 | 12 | 48 | 97 | 294 |
67.01 |
|
7 |
63 | 9 | 4 | 8 | 28 | 37 | 63 | 41.27 |
| 8 | 16 | 2 | 0 | 0 | 0 | 2 | 16 |
87.50 |
|
9 |
9 | 1 | 0 | 0 | 0 | 1 | 9 | 88.89 |
| 10 | 50 | 5 | 0 | 0 | 0 | 5 | 50 |
90.00 |
|
11 |
22 | 2 | 0 | 0 | 0 | 2 | 22 | 90.91 |
| 12 | 24 | 2 | 1 | 1 | 12 | 14 | 24 |
41.67 |
|
17 |
17 | 1 | 0 | 0 | 0 | 1 | 17 | 94.12 |
| 19 | 19 | 1 | 0 | 0 | 0 | 1 | 19 |
94.74 |
|
20 |
40 | 2 | 1 | 2 | 20 | 22 | 40 | 45.00 |
| 21 | 42 | 2 | 0 | 0 | 0 | 2 | 42 |
95.24 |
|
22 |
44 | 2 | 1 | 7 | 22 | 24 | 44 | 45.45 |
| 23 | 46 | 2 | 1 | 1 | 23 | 25 | 46 |
45.65 |
|
Total |
2100 | 536 | 93 | 131 | 372 | 908 | 2100 |
56.76 |
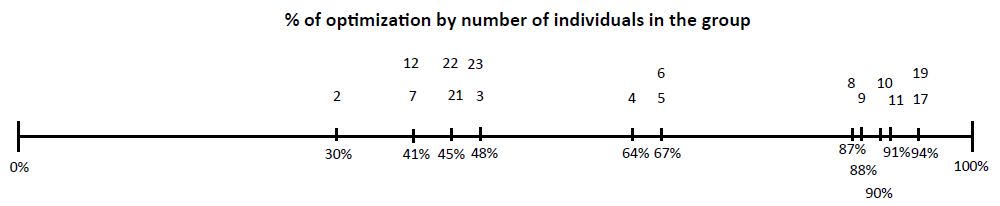
Figure 1: From Left to Right % of optimization below the line. In top of line number of individuals in each group.
Discussion
The SARS COVID-19 pandemic persists till the end of 2020 and probably the first half of 2021, and in the absence of a specific drug and only treatments for the effects of the virus on the coagulation and inflammatory process, the isolation strategy and follow-up of contacts with real-time PCR tests should be followed. The Vaccines probably will modify the number of positive people, but still will be important, given the cost of the tests to be able to optimize resources and group or pool tests is an effective method to do it. Groups of between 4 to 6 people have been the ones with the best optimization results with 64 to 67%, this means that with 1 test it is possible to cover the same number of people and detect more quickly those who must be isolated and follow their contacts. The groups of two are not adequate since an initial test is required and if positive result requires 2 additional tests and this means to carry out for two people three tests. Positive groups of more than 10 people require performing individual tests in subgroups of 2 or 3 and positive ones in individual tests, which is not practical since it takes at least two or three additional days to perform them and although the optimization obtained was of up to 94%, in time and at present time with the expected increase in the number of cases it would not be practical. We found groups of 4 to 6 people are optimal. 3 groups with false positives were detected, these show to start positive above cycle 33 of the 45 cycles of the Rt-PCR programming and the individual tests were always negative. In the groups with cycles less than 31, the positive person or persons were always detected. Two different brands of reagents were used to verify the results in the 3 false positive groups with the same results.
Conclusion
Carrying out group tests in asymptomatic people of open population for any reason, whether it is resuming activities or detecting cases of healthy carriers, is very effective and is optimal in groups 4, 5 and 6 people, this allows more tests to be carried out at a lower cost.
References
- Huipeng Ge, Xiufen Wang, Xiangning Yuan, Gong Xiao, Chengzhi Wang et (2020) The epidemiology and clinical information about COVID-19. European Journal of Clinical Microbiology & Infectious Diseases 39: 1011-1019. [crossref]
- Consultada 21 de octubre 2020
- http://ncov.mohw.go.kr/en/ Tests in Suth Corea updated 1 july 2020
- Ultimo acceso 1 de Agosto 2020
- Chen Y, Liu Q, Guo D (2020) Coronaviruses: genome structure, replication, and pathogenesis Deyin Guo ORCID.
- Zou L, Ruan F, Huang M, Liang L, Huang H et al (2020) SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N engl j med 382: 1171-1179. [crossref]
- Enfermedad por coronavirus, COVID-19 Ministerio de Sanidad España Centro de coordinación de alertas y emergencia sanitaria actualización 4 de abril 2020
- Tapiwa G, Cécile K, Dongxuan Ch, Andrea T (2020) Estimating the generation interval for COVID-19 based on symptom onset medRxiv
- Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S (2020) Virological assessment of hospitalized patients with COVID-2019. Nature 581: 465-469. [crossref]
- Idan Yelin, Noga Aharony, Einat Shaer Tamar, Amir Argoetti, Esther Messer, et (2020) Evaluation of COVID-19 RT-qPCR test in multi-sample pool. medRxiv preprint
- Roni Ben-Ami, Agnes Klochendler, Matan Seidel, Tal Sido, Ori Gurel-Gurevich, et (2020) Large-scale implementation of pooled RNA extraction and RT-PCR for SARS- CoV-2 detection. Clinical Microbiology and Infection PII: S1198-743X(20)30349-9.
- Michael G Becker, Tracy Taylor, Sandra Kiazyk, Dana R Cabiles, Adrienne FA Meyers, et al. (2020) Recommendations for sample pooling on the Cepheid 1 GeneXpert® system using the Cepheid Xpert® Xpress SARS-2 CoV-2 bioRxiv preprint
- Abdalhamid B, Bilder CR, McCutchen EL, Hinrichs SH, Koepsell SA, etal. (2020) Assessment of Specimen Pooling to Conserve SARS CoV-2 Testing Am J Clin Pathol 153: 715-718. [crossref]
- Ola Brynildsrud (2020) COVID-19 prevalence estimation by random sampling in population – optimal sample pooling under varying assumptions about true prevalence BMC Med Res Methodol 20: 196.