Abstract
Ordered DNA methyltransferases covalently bound to Y-junction DNA were spontaneously taken up mammalian cells in culture, where a nuclear localization signal carried by the bound methyltransferases caused the fluorescently labeled nucleoprotein complex to decorate the nucleus of several mammalian cell lines. We report proof of concept experiments demonstrating that the Y-junction nucleoprotein assembly can be modified to carry gene expression cassettes that permit spontaneous transfection and expression of a desired protein in mammalian cells.
Introduction
Ordered nucleoprotein assemblies have been developed for a variety of applications in Nanofabrication [1]. Many protein bearing nanoparticles have been shown to target mammalian cells where they can be selectively internalized [2,3]. Bacterial DNA methyltransferases covalently bound to Y-junction DNA have also been shown to be spontaneously taken up by mammalian cells in culture [4]. Further, those experiments showed that the nuclear localization signal carried by the bound methyltransferases caused the fluorescently labeled nucleoprotein complex to decorate the nucleus of several mammalian cell lines. Here we report proof of concept experiments using Green Fluorescent Protein (GFP) expression to demonstrate that the Y-junction nucleoprotein assembly can be modified to carry gene expression cassettes that permit spontaneous transfection and expression of a desired protein in mammalian cells.
Materials and Methods
Cell Culture conditions, EcoRII DNA methyltransferase purification, flurocytidine containing DNA Y-junction synthesis and construction, as well as the construction of the thioredoxin-linked nanoparticle have all been described in detail [4–6]. The Y-Junction was modified for expression cassette delivery as described in figure 1.
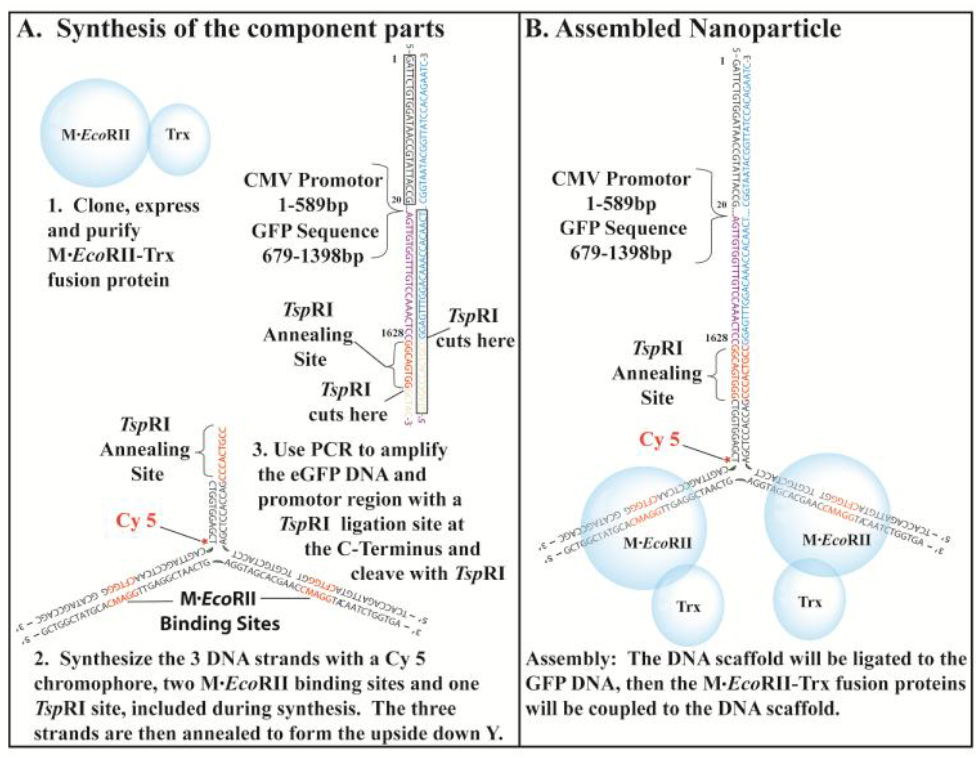
Figure 1. The NP-Trx2-GFP-DNA was assembled from the component parts.
Synthesis of the component parts. 1. The EcoRII Methyltransferase fusion protein was cloned, expressed and purified from E. coli. 2. The DNA scaffold is composed of three DNA strands with a Cy 5 tag added during the synthesis. The sequences for two M.EcoRII binding sites and one Tsp RI Ligation site are designed into the sequences. 3. The GFP DNA was generated by PCR, which will include the GFP open reading frame and the eukaryotic promoter sequence. A Tsp RI sequence was included in the N-terminal region of the GFP DNA. B. The assembly consists of three steps. First the three strands of the DNA scaffold were annealed. Then the GFP DNA was ligated to the DNA scaffold. Finally the EcoRII Methyltransferase fusion proteins were coupled to the EcoRII Methyltransferase binding sites.
Transfection Conditions: Comparison of NP-Trx2-GFP -DNA Mediated GFP Delivery with Lipid Mediated Transfection.
We compared the effectiveness of delivering NP-Trx-GFP-DNA to DU145 cells with a traditional transfection reagent (e.g. Lipofectamine™ (Invitrogen)). Cells were plated at 5,000 cells per plate (Costar) before delivery without antibiotics and grown to 50–70% confluence. The media was removed from each well and the cells were washed with PBS. Then 1.0 µM NP-Trx2-GFP-DNA was added to the cells and incubated for multiple time points at 37oC in 5% CO2. The transfected cells were observed by fluorescent microscopy and fluorimetry for the frequency of green fluorescent protein expression in viable cells per input viable cell. The emission wavelength of Cy5.5 at 707nm, which is on the DNA Y-Junction (Figure1), did not interfere with the appearance of fluorescence from GFP at 509nm. Following incubation, the cells were washed with PBS and growth media was added and the cells were incubated to permit growth. The cells were monitored using an inverted fluorescent microscope over the next 10 days for internalization at 707nm and for the appearance of GFP fluorescence at 509nm indicating successful expression of GFP.
Results and Discussion
As noted above the trisubstituted nanoparticle was found to spontaneously enter mammalian cell lines of several types (Figure 2). Among those, the DU145 cell line, which showed significant uptake in the nucleus and HK293, which showed weak uptake were selected for further work in this proof of concept report.
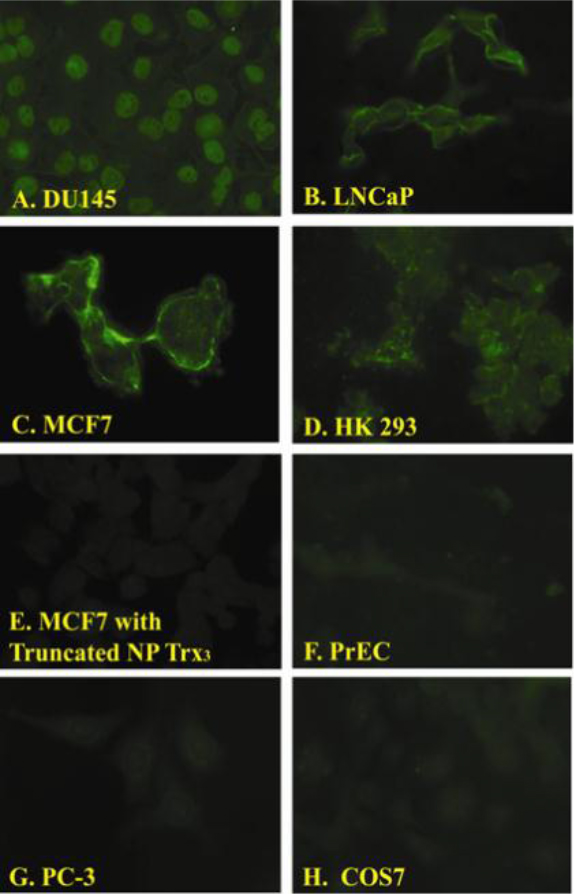
Figure 2. Fluorescent images of cell lines exposed to the NP-Trx3.
We observed differential binding of the NP-Trx3 prepared as described in [4] with different cell lines. Fluorescence was observed in DU145 (Panel A), LNCaP (Panel B), MCF-7 (Panel C), HK293 (Panel D) but not MCF7cells exposed to Y-Junction DNA without the targeting ligand (Eco RII DNA methyltransferase) (Panel E), Primary Prostate Epithelial Cells (PrEC) (Panel F), PC-3 (Panel G) and COS7.
Final assembly of the carrier nanoparticle (NP-Trx2-GFP-DNA) was carried out as described in materials and methods. Each stage in the process was tested using microfluidics mobility shift analysis [7]. The results of a typical mobility shift analysis are depicted in Figure 3.Transfection efficiency was monitored by GFP fluorescence after transfection. Nuclear fluorescence indicating GFP expression was observed with DU145 cells using Y-GFP DNA and Lipofectamine mediated transfection or with purified NP-Trx2-Y-GFP-DNA without Lipofectamine mediation. The mixture of unreacted components of the nanoparticle (Y-DNA, GFP-DNA, and EcoRII-Trx) did not yield GFP expression (Figure 4). Unpurified Np-Trxn-Y-GFP-DNA was found to exhibit GFP expression without Lipofectamine mediation suggesting that the nanopaticle might support transfection with a single EcoRII-Trx molecule acting as targeting ligand. HK293 Cells were also found to support NP-Trx2-Y-GFP-DNA without Lipofectamine mediation although with lower efficiency than DU145 cells.
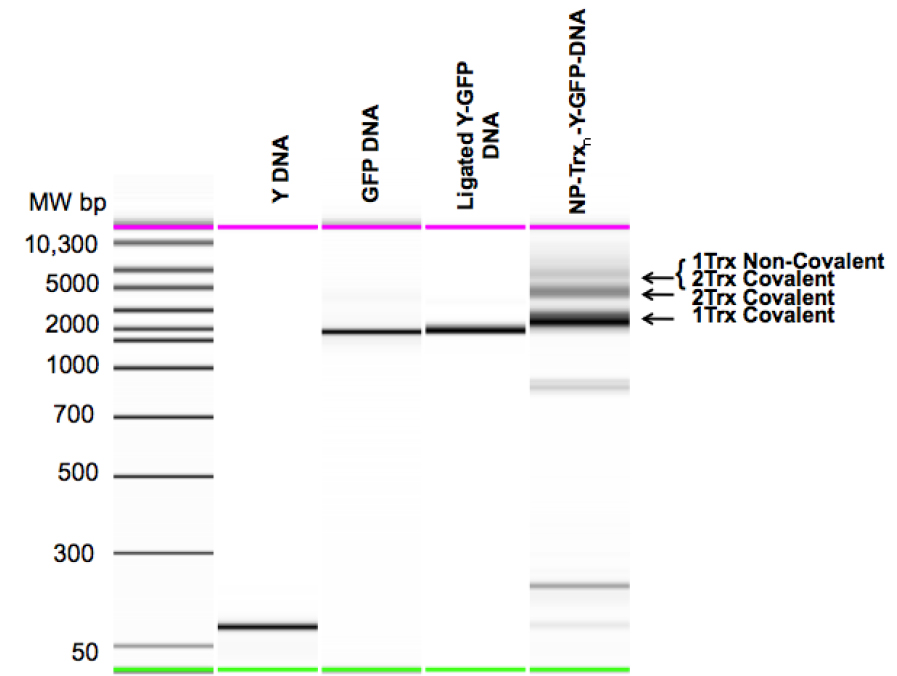
Figure 3. Mobility Shift analysis of the Steps in Carrier Nanoparticle Assembly.
Y DNA: Purified Y-Junction. GFP DNA: Purified TspR1 cleaved GFP expression cassette. Ligated Y-GFP DNA: Purified Y-DNA GFP DNA ligation product. NP-Trxn-Y-GFP-DNA: Unpurified EcoRII-Txn + Y-GFP reaction products, showing DNA carrying one or two covalently bound EcoRII-Txn molecules and two covalently bound and one non-covalently bound EcoRII-Txn molecules.
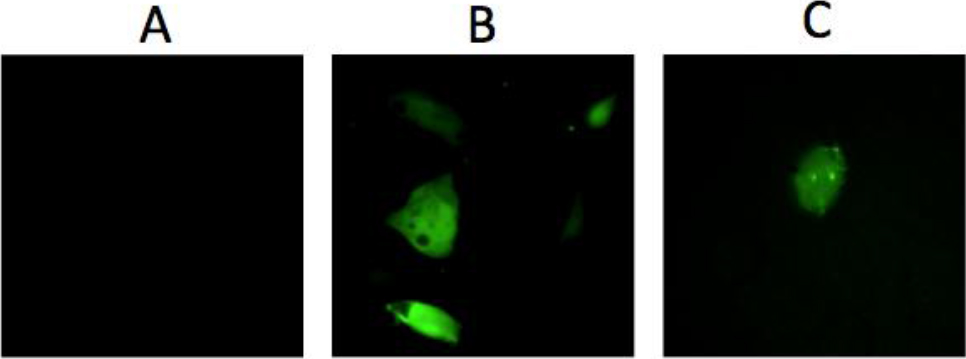
Figure 4. GFP Expression after transfection.
A: Lack of GFP expression after exposing DU145 cells to the mixture unreacted components of the nanoparticle (Y-DNA, GFP-DNA, and EcoRII-Trx). B: GFP expression after Lipofectamine mediated transfection of DU145 cells with 300nM Y-GFP-DNA. C: GFP expression after exposing DU145 cells to 300nM purified NP-Trx2-Y-GFP-DNA alone.
Expression was seen to be transient with the number of fluorescent nuclei diminishing steadily over a period of ten days, suggesting that the DNA from the nanoparticle did not become integrated. Transfection efficiency was comparable to that observed with Lipofectamine mediation transfection, and depended on the cell line used for transfection.
For DU145 cells 38% transfection was observed with Nanoparticle mediated or Lipofectamine mediated transfection assuming the input transfectant was not replicated. However, the efficiency was adequate for most purposes and avoids the undesirable cellular stress of Lipofectamine exposure [8]. We also noted that purification of the NP-Trx2-Y-GFP-DNA DNA was unnecessary since simply allowing the reaction between the Y-GFP-DNA and EcoRII-Trx to occur in 30 min incubation at 37oC resulted in comparable transfection levels. We were not concerned that uncoupled EcoRII activity might alter gene expression by methylating non CG methylation sites since previous work with this enzyme showed that this does not occur [9]. Further, given the ability of the of the nanoparticle to spontaneously enter the cytoplasm of certain cells (e.g. LNCaP see Figure 2B) the system can be modified to carry siRNA by repurposing the truncated Y-Junction so as to permit hybridization of a double stranded siRNA molecule to the nucleoprotein complex, thereby avoiding the deleterious effects of the Lipofectamine transfection generally employed in siRNA transfection [8].
References
- M.Niemeyer C (2010) Halbsynthetische DNA-Protein-Konjugate fur Biosensorik und Nanofabrikation. Angewandte Chemie 122: 1220–1238.
- Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, et al. (2004) Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol 22: 198–203.
- Mortensen MW, Bjorkdahl O, Sorensen PG, Hansen T, Jensen MR, et al. (2006) Functionalization and cellular uptake of boron carbide nanoparticles. The first step toward T cell-guided boron neutron capture therapy. Bioconjug Chem 17: 284–290.
- Singer EM, Crocitto LE, Weiss LM, Loera S, Imam SA, et al. (2011) Biomarker Identification with Ligand-Targeted Nucleoprotein Assemblies Nanomedicine (London) 6: 659–668.
- Clark J, Singer EM, Korns DR, Smith SS (2004) Design and analysis of nanoscale bioassemblies. Biotechniques 36: 992–996, 998–1001.
- Singer EM, Smith SS (2006) Nucleoprotein assemblies for cellular biomarker detection. Nano Lett 6: 1184–1189.
- Clark J, Shevchuk T, Swiderski PM, Dabur R, Crocitto LE, et al. (2003) Mobility-shift analysis with microfluidics chips. Biotechniques 35: 548–554.
- Dalby B, Cates S, Harris A, Ohki EC, Tilkins ML, et al. (2004) Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods 33: 95–103.
- Shevchuk T, Kretzner L, Munson K, Axume J, Clark J, et al. (2005) Transgene-induced CCWGG methylation does not alter CG methylation patterning in human kidney cells. Nucleic Acids Res 33: 6124–6136.