Abstract
A clinically ill red-eyed tree frog (Agalychnis callidryas) was submitted to the Escuela de Medicina Veterinaria, Universidad Nacional, Heredia, Costa Rica that was infested with slender subcutaneous parasites located in its dorsal subcutis. We humanely euthanized the frog and the parasites and tissues collected for further study. Light microscopic examination of histological sections of the frog’s heart and stomach displayed numerous microfilaria in these tissues. DNA was isolated from the adult nematodes and PCR used to amplify regions of the 18S small ribosomal subunit (18S rRNA), 28S large ribosomal subunit (28S rRNA), mitochondrial cytochrome oxidase 1 (COI) gene and the mitochondrial 12S ribosomal subunit (12S rRNA). The amplicon DNA sequences were determined, and submitted as BLAST searches of the NIH GenBank nucleotide database. Results demonstrated that portions of the parasites gene sequences were unique, but closely related to nematodes in the superfamily Filarioidea. The 4 gene sequences of the red-eyed frog parasite gene sequences were concatenated and aligned with concatenated sequences of the same 4 gene regions in 35 other species within the superfamily Filarioidea, and 1 species in the superfamily Spirurida as the outgroup, for phylogenetic analysis using MEGA X software. We aligned the dataset using MUSCLE, analyzed for the evolutionary model that best fit the data using jModeltest, followed by tree construction using a Maximum Likelihood method of phylogenetic analysis. The results assign the filarial parasite of the red-eyed tree frog to the genus Ochoterenella. DNA isolated from the adult parasites did not contain 16S rRNA sequences of the bacterium Wolbachia, consistent with other members of the Ochoterenella genus. Based on our phylogenetic analysis of the concatenated 4 gene sequences from this parasite, review of the current literature, and the subcutaneous location of the adult parasites in the frog, we surmise this is the first molecular characterization of this filarial parasite of the red-eyed tree frog.
Keywords
Agalychnis calidryas, Costa Rica, Filarioidea, Microfilaria, Phylogeny, Wolbachia
Introduction
Nematodes of the superfamily Filarioidea consist of parasites of vertebrate animals some of which are associated with pathology in humans and animals [1]. The adult filarid parasites dwell in body cavities, blood vessels, lymphatic vessels, subcutaneous tissues or the eye depending on the species. Female filarial parasites produce microfilaria offspring that circulate in the blood, lymphatic tissues and tissue fluids. Microfilaria ingested by biting arthropods that feed on host species blood, lymph and/or subcutaneous tissues fluids further develop to an infectious stage transmitted in subsequent blood meals. Biting arthropods are an obligatory intermediate host in the life cycle of filarial parasites and once ingested the microfilaria molt continuing their development. Moreover, the biological relationship between some genera of filarial parasites and arthropods may have included transfer of the endosymbiotic bacteria in the genus Wolbachia, most commonly found in the gametes of arthropods, particularly insects, but also found in some filarial genera [2].
Wolbachia are bacterial endosymbionts that provide energy rich metabolites to their host cells similar to the role mitochondria play in eukaryotic cells [2]. In the relationship with filarial hosts, Wolbachia supply energy supporting metabolically demanding stages of the filarid’s life such as production of microfilaria. Wolbachia likely co-evolved with some filariae from a single infection event and their removal sterilizes dependent female filariae species [3].
During routine surveillance of native frogs in Costa Rica to assess their blood for the presence of hematogenous parasites, a single red-eyed tree frog (Agalychnis callidryas) was captured that was infested with slender round parasites in the subcutis of over the dorsal lymph sacs. The subcutaneous nematodes were isolated and DNA sequences of four genes were determined and analyzed using standard molecular methods. Through the molecular analyses of gene sequences in this study, and review of the literature, we determined that the red-eyed tree frog filarial parasite is in the genus Ochoterenella had not previously been characterized.
Materials and Methods
Specimen Collection
A clinically ill frog Agalychnis callidryas from the province of Guanacaste, Costa Rica, was referred to the Parasitology laboratory, and given case number PA-043-19. The frog was euthanized according to the current AVMA guidelines for euthanasia of animals [4]; benzocaine was topically applied to the inguinal area of amphibian, and once immobilized from the drug it was placed in refrigeration for 30 minutes. Four adult nematodes were found in the dorsal subcutaneous tissues and two were used for this molecular sequence analysis. Tissues and organs of the frog were collected post-mortem and fixed in 10% buffered formalin solution overnight for histopathologic examination. Paraffin-embedded sections (five μm) were cut and stained with hematoxylin and eosin (H&E). Two additional five micrón paraffin sections of the frog’s heart and stomach containing microfilaria were collected for DNA isolation. The parasites were not adequately preserved to obtain morphological measurements, and were dehydrated in 100% ethanol prior to isolating their DNA.
Gene Amplification and Cloning
DNA was isolated from two adult filarial parasites using DNeasy Tissue Kit (Qiagen®, Germantown, Maryland) by macerating the nematodes in a one-ml glass tissue grinder containing 180µL of ATL buffer and 20 µL proteinase-K. The proteinase-K digestion proceeded overnight at 55°C. DNA isolation proceeded the next day according to manufacturer’s recommendations for animal tissue, we used 50 µL of 70°C buffer AE for the final DNA elution. DNA was isolated from paraffin sections by dissolving two (five µM) sections in xylene overnight, followed by sequential one hr rehydration steps in 100% ethanol, 70% ethanol followed by water. Digestion of the de-paraffinized tissue in ATL buffer and proteinase-K at 55°C proceeded overnight and DNA was isolated using DNeasy Tissue Kit (Qiagen) according to manufacturer’s recommendations, however DNA elution used 50 µL of 70°C buffer AE.
Isolated nematode DNA was quantified on a ThermoFisher® Nanodrop Lite spectrophotometer (ThermoFisher, Wilmington, Delaware) and two µL samples were subjected to five different PCR reactions using Platinum Taq polymerase (Thermofisher-Invitrogen®, Carlsbad, California): 12S rRNA, 18S rRNA, 28S rRNA, COI and Wolbachia 16S rRNA.
The PCR primer sequences used to amplify the nematode 18S rRNA have been previously described [5]. The nematode cox1 PCR primer sequences were designed from a MUSCLE alignment created using MEGA X software Kumar et al. (2018) analysis of GenBank accessions of COI in Loa loa (AJ544875), Dirofilaria repens (AB973225), Brugia malayi (KP760171), Setaria digitata (EF174427) and Diptelonema evansi (KR184816). The PCR primer sequences used targeting the 12S rRNA and 28S rRNA genes were those published [6-8]. We used the primer sequences that targeting the 16S rRNA of Wolbachia bacteria published [3]. DNA sequences for all primers and primer annealing conditions for the five amplification reactions appear in Table 1.
Table 1: PCR Primer sequences and annealing conditions.
|
Primer Designation |
Primer Sequence (5’-3’) | Target |
Annealing (°C) |
| Filarid mMCO1F |
GTAGTTGAACTTTTTAYCCTCC |
COI |
55 |
| Filarid mMCO1R |
AACAGCAATYCARATAGAAGCAA |
||
| Nema 18S F635 |
GAGGGCAAGTCTGGTGCCAGCAG |
18S rDNA |
65 |
| Nema 18S R1728 |
YATACCTATTCGAAGGGATAG |
||
| 12SF |
GTTCCAGAATAATCGGCTA |
12S rDNA |
50 |
| 12SdegR |
ATTGACGGATGRTTTGTACC |
||
| F28SF1 |
CCTCAACTCAGTCGTGATTACC |
28S rDNA |
58 |
| F28SintdR1* |
TCTTYACTTTCATTAYGCTT |
||
| Wolbachia 16SF |
YATACCTATTCGAAGGGATAG |
16S rDNA |
45 |
| Wolbachia 16SR |
AGCTTCGAGTGAAACCAATCC |
||
Extension for all reactions was at 72°F for one minute/kilobase, 40 PCR cycles.
We cloned one microliter of each PCR amplicon into plasmid pCR4-TOPO (Thermofisher-Invitrogen) and used to transform chemically competent TOP-10 Escherichia coli (Thermofisher-Invitrogen). The transformed TOP-10 bacteria were grown overnight on Luria-Bertani agar containing kanamycin 50µg/mL (LBK agar). We picked six clones the next day and inoculated into individual six mL LBK broth cultures, and were grown overnight. The pCR4 plasmid containing amplicon insert was isolated from each clone’s broth culture using Plasmid Miniprep (Qiagen) and the purified plasmid DNA diluted to 50 ng/µl in 2 mM EDTA buffer. Plasmid clones were sent to Genewiz, LLC (South Plainfield, New Jersey) and the amplicon nucleic acid sequences determined by the Sanger method, initiating sequencing from both of the T3 and T7 promoter sites located upstream of the amplicon on opposite DNA strands. We analyzed the sequences obtained using the software suite MEGA X [6]. Plasmid sequences were removed from the resulting forward and reverse amplicon sequences, 1 strand from each clone was reverse transcribed, and the amplicon information from all clones were aligned using the MUSCLE algorithm to obtain a consensus sequence for each of the 4 genes from the red-eyed tree frog filarial parasite.
Phylogenetic Analyses
The NIH GenBank accession for all four genes of the red-eyed tree frog filarial parasite are in Table 2. The GenBank accession information for the homologous gene sequences of the other 35 filarial parasites and one outgroup used for phylogenetic analysis are provided in Table 2. All manipulation of DNA sequences used the software package MEGA X.
Table 2: Species within the superfamily Filarioidea in the analysis, rooted to a member of superfamily Spiruridea
|
Organism |
cox1 |
12S | 18S |
28S |
| Ochoterenella sp. 1 SHF-2019 |
MN368875 |
MT150113 | MN334554 |
MT153694 |
| Acanthocheilonema viteae |
KP760169 |
KX022983 | KP760117 |
KP760359 |
| Breinlia jittapalapongi |
KP760170 |
KP760316 | KP760119 |
KP760361 |
| Brugia pahangi |
MT027204 |
KP760318 | KP760121 |
KP760363 |
| Brugia timori |
KP760173 |
KP760319 | KP760122 |
KP760364 |
| Cercopithifilaria bainae |
KP760175 |
KP760321 | KP760123 |
KP760365 |
| Cruorifilaria tuberocauda |
KP760176 |
KP760322 | KP760125 |
KP760367 |
| Dipetalonema caudispina |
KP760178 |
KP760323 | KP760127 |
KP760369 |
| Dipetalonema gracile |
KP760181 |
KP760326 | KP760130 |
KP760372 |
| Dipetalonema graciliformis |
KP760182 |
KP760328 | KP760131 |
KP760373 |
| Dipetalonema robini |
KP760183 |
KP760329 | KP760132 |
KP760374 |
| Dirofilaria immitis |
KT716014 |
KP760330 | KP760133 |
KP760375 |
| Dirofilaria repens |
KP760185 |
KP760331 | KP760134 |
KP760376 |
| Foleyella candezei |
KP760187 |
FR827906 | KP760136 |
KP760378 |
| Icosiella neglecta |
KP760189 |
KP760334 | KP760138 |
KP760380 |
| Litomosoides brasiliensis |
KP760191 |
KP760336 | KP760140 |
KP760382 |
| Litomosoides hamletti |
KP760192 |
KP760337 | KP760141 |
KP760383 |
| Litomosoides solarii |
KP760193 |
KP760338 | KP760142 |
KP760385 |
| Loa loa |
KP760194 |
KP760339 | KP760143 |
KP760386 |
| Loxodontofilaria caprini |
AM749242 |
AM779822 | KP760144 |
KP760387 |
| Madathamugadia hiepei |
JQ888272 |
JQ888290 | KP760146 |
KP760389 |
| Mansonella ozzardi |
KP760195 |
KP760340 | KP760147 |
KP760390 |
| Monanema martini |
KP760196 |
KP760341 | KP760149 |
KP760391 |
| Ochoterenella sp. 1 EL-2015 |
KP760198 |
KP760343 | KP760151 |
KP760394 |
| Ochoterenella sp. 2 EL-2015 |
KP760199 |
KP760344 | KP760152 |
KP760395 |
| Ochoterenella sp. 3 EL-2015 |
KP760197 |
KP760342 | KP760150 |
KP760393 |
| Onchocerca dewittei japonica |
KP760203 |
KP760349 | KP760154 |
KP760397 |
| Onchocerca gutturosa |
AJ271617 |
KP760347 | KP760156 |
KP760399 |
| Onchocerca ochengi |
KC167358 |
KP760348 | KP760157 |
KP760400 |
| Onchocerca skrjabini |
AM749274 |
AM779809 | KP760158 |
KP760401 |
| Oswaldofilaria chabaudi |
KP760204 |
KP760350 | KP760159 |
KP760402 |
| Oswaldofilaria petersi |
KP760205 |
KP760351 | KP760160 |
KP760403 |
| Pelecitus fulicaeatrae |
KP760206 |
KP760352 | KP760161 |
KP760404 |
| Protospirura muricola |
KP760207 |
KP760353 | KP760162 |
KP760405 |
| Rumenfilaria andersoni |
JQ888279 |
JQ888297 | KP760163 |
KP760406 |
| Setaria labiatopapillosa |
MF589585 |
KP760354 | KP760164 |
KP760407 |
| Setaria tundra |
KU508985 |
KP760355 | KP760165 |
KP760408 |
Individual MUSCLE alignments (in MEGA X) were created for each of the 4 genes using sequences from our red-eyed tree frog filarial parasite, and the homologous gene sequences from 35 Filarioidea and 1 Spirurida outgroup (Protospirurida muricola). We truncated each gene sequence so that all the alignment begins at the same 5’-nucleotide position (with the one exception of the 5’ end of COI gene of Monanema martini) and end at the same 3’ nucleotide position. These alignments were concatenated (using MEGA X) forming a 2,604 nucleotide long dataset, and aligned with the MUSCLE (non-coding) algorithm. The best evolutionary model for the concatenated dataset was determined using jModeltest [7]. Phylogenetic analysis was performed using the maximum likelihood method with the following settings: 1,000 bootstrap replicates, GTR+G+I model, six discrete gamma transition/transversion rates, Nearest-Neighbor-Interchange heuristic method of tree inference, and the branch swap filter set at moderate. The resulting phylogram was rooted to the Spirurida outgroup, nodes with less than 50% bootstrap agreement were collapsed and the phylogram exported for text annotations using Corel Draw® (Ottawa, Ontario, Canada).
Results
Figure 1 is a photograph of the live restrained Agalychnis callidryas prior to euthanasia and necropsy. The dorsal skin visibly deformed was due to the presence of adult nematodes in the subcutis. Light microscopic examination of H&E stained sections of heart and stomach revealed microfilaria in the small vessels of the heart and stomach (Figure 2a and 2b).
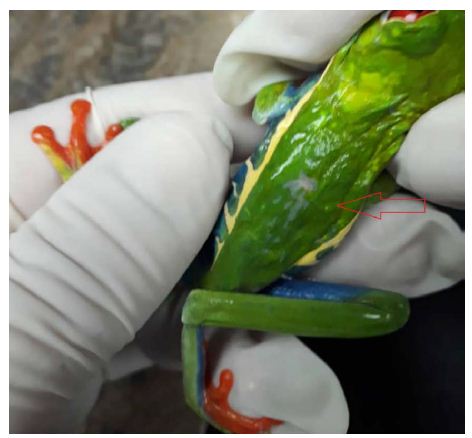
Figure 1: Depicted is a photograph of the restrained red-eyed tree frog (Agalychnis callidryas), a nematode is located in the subcutis seen at the tip of the arrow.
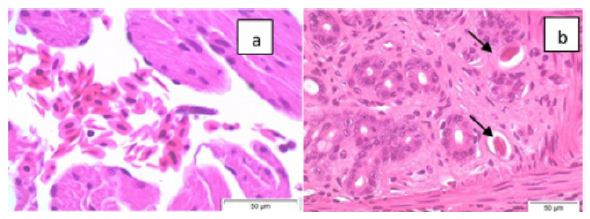
Figure 2: Photomicrographs (x60 magnification) of H&E stained paraffin-embedded sections of heart (a) and stomach (b) showing microfilaria (arrows) in these tissues.
Gene specific PCRs amplified 1,098 bp of the 18S rRNA, 1,131 bp of the 28S rRNA, 470 bp of COI gene and 503 bp of the 12S rRNA from the red-eyed tree frog filarial parasite. We deposited the sequences for each gene from this parasite of the red-eyed frog into the NIH GenBank (accession numbers appear in Table 2). BLAST search of the GenBank nucleotide database using each gene sequence from the red-eyed tree frog filarial parasite as the subject, and the BLAST default search settings, retrieved members of the superfamily Filarioidea. Moreoever, the red-eyed tree frog parasite has the highest degree of similarity to sequences of members in the genus Ochoterenella. Sequence identity between the concatenated sequence of the red-eyed tree frog Ochotenerella and the other Ochotenerella sequences from the GenBank are in Table 3. The concatenated gene sequences of the red-eyed tree frog Ochotenerella has 96.7% identity with Ochotenerella sp. 3 EL-2015 (Table 3).
Table 3: Pairwise similarities between concatenated sequences of Ochotenerella species.
|
Ochotenerella sp. 1 SHF-2019 |
Ochotenerella sp. 3 EL-2015 | Ochotenerella sp. 1 EL-2015 |
Ochotenerella sp. 2 EL-2015 |
|
| Ochotenerella sp. 1 SHF-2019 |
100 |
|||
| Ochotenerella sp. 3 EL-2015 |
96.7 |
100 |
||
| Ochotenerella sp. 1 EL-2015 |
92.1 |
91.5 |
100 |
|
| Ochotenerella sp. 2 EL-2015 |
92.1 |
92.7 | 95.1 |
100 |
DNA isolated from paraffin sections of heart and stomach subjected to COI PCR produced amplicons whose DNA sequence was identical to that of the COI sequence from the adult filarid in the subcutis.
jModeltest analysis determined that the best fit evolutionary model for our nucleotide dataset is General Time Reversible, with six gamma distributed rates, and some invariant sites (GTR+G+I). GTR+G+I had the lowest corrected Akaike Information Criteria and Bayesian Information Criteria when compared to 88 other evolutionary models in this analysis. The jModeltest tree using the GTR+G+I model with the highest log likelihood had a value of -23584.41, a rate Gamma distribution with six categories (+G, parameter = 0.2963) and 26.32% of sites evolutionarily invariable. Tree construction using maximum likelihood with bootstrap phylogenetic analysis grouped the red-eyed tree frog filarial parasite with members in the genus Ochoterenella, with other filaria known to parasitize frogs in the Central and South Americas (Figure 3). The red-eyed tree frog Ochotenerella is most closely related to Ochoterenella sp. 3 EL-2015 voucher 194JW MNHN, which parasitizes Phyllomedusa bicolor the Brazilian tree frog (also called blue-and-yellow frog, bi-colored tree frog, giant monkey frog, giant-leaf frog, or waxy-monkey tree frog) in the Family Hylidae. The red-eyed tree frog Ochoterenella and Ochoterenella sp. 3 EL-2015 form a subclade with two other species of Ochoterenella, the latter nematodes parasitizing anurans in the family Bufonidae: Rhinella marina (the cane toad; Ochoterenella sp. 2 EL-2015 voucher 194JW MNHN) and Rhinella granulosa (the granular toad, common lesser toad; Ochoterenella sp. 1 EL-2015 voucher 194JW MNHN).
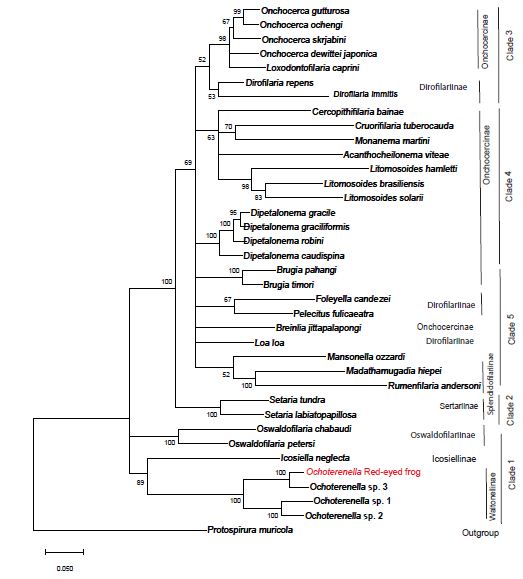
Figure 3: The phylogenetic tree represents the evolutionary history inferred by using the maximum likelihood method using the General Time Reversible model. The tree with the highest log likelihood (-23746.39) is shown. The percentage of trees in which the taxa group together is next to the branches points (based on consensus among 1,000 replicates). Partitions in which the percentage of trees is less than 50% bootstrap replicates are collapsed, partitions 50% consensus or greater are shown next to the branches. The length of each branch corresponds to the number of nucleotide substitutions per site and we provide a scale for branch length. Depicted on the right are the eight traditional subfamilies determined by morphological characters and to their right are the 5 ONC clades proposed by Lefoulon et al. (2015).
Wolbachia PCR of DNA isolated from the adult red-eyed tree frog filarial parasite did not produce a 450 bp amplicon, when compared with the amplicon resulting from PCR of total mosquito DNA as a positive control (data not shown).
Based upon our analysis of the concatenated gene sequences of the red-eyed frog filarid, the host Agalychnis callidryas, the parasite’s unique anatomical location in the host, and the absence of Wolbachia, we determined that this parasite is undescribed previously by nucleic sequence analysis and represents a unique molecular taxonomic unit.
Discussion
This is the initial molecular characterization of a red-eyed tree frog subcutaneous filarial parasite, which according to our analysis is in the genus Ochoterenella. Our data and analyses recapitulate a portion of the data from a more detailed multi-locus study of Filarioidea published by Lefoulon [8]. The study by Lefoulon [8] used sequences from three additional gene loci (hsp70, Rbp1 and myoHC) and included 11 additional filarial species in their analysis, beyond the four loci and 36 species in the current study. In that previous study and our current study, both datasets supported the GTR+G+I evolutionary substitution model. In the previous study by Lefoulon [8], the authors concluded that the 46 members of the superfamily Filarioidea in their study should be subdivided into five clades (designated ONC1 through ONC 5), not the eight subfamilies previously created using morphological characters. The ancestral clade is ONC1, containing members of the genera Oswaldofilaria, Icosiella and Ochoterenella), the ONC2 diverged from ONC1 and contains members of the genus Setaria, and the clade ONC3 contains Onchocerca, Loxodontofilaria and Dirofilaria. Our current study supports the conclusion of Lefoulon [8] to assign those same genera to the subfamilies ONC1, ONC2 and ONC3 abandoning the previous subfamily nomenclature. However, the further grouping by Lefoulon [8] of two additional clades (ONC4 and ONC5) is unsupported by our analysis. Comparing our study to that of Lefoulon [8], our study lacks the sequence information from three additional genes (myoHC, Rbp1, Hsp70). The additional information from three genes resulted in better resolution of relationships that supported Lefoulon [8] separating the ONC4 and ONC5 clades. Our molecular data supports the conclusion that the red-eyed frog filarid parasite had not previously characterized by molecular methods, and that this parasite is in the genus Ochoterenella whose members parasitize frogs. Of the four Ochoterenella that have been characterized by molecular analyses, the two that parasitize Hylidae (tree frogs) show greater similarity to each other relative to the two Ochotenerella that parasitize Bufonidae (true toads).
Previous surveys of nematode parasites in Hyalid anurans in Area de Conservacion Guanacaste, Costa Rica did not detect any microfilaria in their blood [9,10]. The Checklist of Helminth parasites of Amphibians from South America [11] catalogs publications of Filarioidea forms in Hyaloidea none of which include location of adult parasites in the subcutaneous tissues of their host: Foleyella convoluta in the body cavity of Hypsiboas faber, Leptodactylus latrans, and Leptodactylus pentadactylus; Ochoterenella convoluta in the body cavity or intestines of Dendropsophus microcephalus (Hyla microcephala), Scinax nebulosus, Leptodactylus fuscus (Leptodactylus silbilatrix and Leptodactylus typhonius), Leptodactylus latrans and Leptodactylus pentadactylus; Ochoterenella digicaudata in the body cavity of Hypsiboas albopunctata, Hypsiboas lanciformis, Leptodactylus labyrinthicus, Leptodactylus latrans, Trachycephalus mesophaeus and Hyla mesophaea; Ochoterenella scalaris in sublingual tissue and body cavity of Leptodactylus latrans and Leptodactylus pustulatus; and Ochoterenella vellardi the body cavity of Osteocephalus taurinus, Hypsiboas (Boana) fasciatus (Hyla fasciata), and Osteocephalus taurinus.
The arthropod intermediate host that transmits the red-eyed frog Ochoterenella is unknown. The intermediate host for the life cycle of most filaria of frogs are either ticks or mites, although mosquitos could also function in this role. Determining the intermediate host of the red-eyed tree frog filarial parasite will provide insight into the geographic range of amphibian hosts that may harbor this nematode. Studies that included examining filarial parasites of amphibians and reptiles for Wolbachia [2], concluded that members of the genus Ochoterenella did not contain the endosymbiont bacteria, recapitulated by our finding in the filarid of the red-eyed tree frog.
Abbreviations
DNA: Deoxyribonucleic Acid
16S rRNA: Bacterial Small Ribosomal Subunit Gene
18S rRNA: Eukaryotic Large Ribosomal Subunit Gene
12S rRNA: Mitochondrial Small Ribosomal Subunit Gene
28S rRNA: Eukaryotic Large Ribosomal Subunit Gene
COI: Mitochondrial Cytochrome Oxidase Type I Gene
myoHC: Myosin Heavy Chain Gene
Rbp1: DNA-Dependent RNA Polymerase Type 1 Gene
Hsp70: Heat-Shock Protein 70 Kilodalton Gene
µL: Microliter
µM: Micromolar
LBK: Luria-Bertani Agar or Broth with kanamycin.
References
- Eamsobhana P, Lim PE, Yong HSc (2013) Molecular phylogeny of filarial worms (Nematoda: Filarioidea). The Raffles Bulletin of Zoology 29: 99-109.
- Taylor MJ, Voronin D, Johnston KL, Ford L (2013) Wolbachia filarial interactions. Cellular Microbiology 15: 520-526. [crossref]
- Ferri E, Bain O, Barbuto M, Martin C, Lo N, et al. (2011) New insights into the evolution of Wolbachia infections in filarial nematodes inferred from a large range of screened species. PLOS One 6: 20843. [crossref]
- Leary S, Underwood W, Anthony R, Cartner S, Grandin T, et al. (2020) AVMA Guidelines for the Euthanasia of Animals: 2020 Edition. American Veterinary Medical Association, Schaumburg, Illinois.
- Feldman SH, Bowman SG (2007) Molecular phylogeny of the pinworms of mice, rats and rabbits, and its use to develop molecular beacon assays for the detection of pinworms in mice. Lab Animal (Nature) 36: 43-50. [crossref]
- Kumar S, Stecher G, Mi L, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol and Evol 35: 1547-1549. [crossref]
- Posada D (2008) jModelTest: Phylogenetic model averaging. Mol Biol and Evol 25: 1253-1256. [crossref]
- Lefoulon E, Bain O, Bourret J, Junker K, Guerrero R, et al. (2015) Shaking the tree: Multi-locus sequence typing usurps current Onchocercid (filarial nematode) phylogeny. PLoS Neglected Trop Dis 9: 0004233. [crossref]
- Bursey CR, Brooks DR (2010) Nematode parasites of 41 anuran species from the Area de Conservacion Guanacaste, Costa Rica. Comp Parasitol 77: 221-231.
- Desser SS (2001) The blood parasites of anurans from Costa Rica with reflections on the taxonomy of their trypanosomes. J Parasitol 87: 152-160. [crossref]
- Compião KM, Morais DH, Dias OT, Aguiar A, Toledo G, et al. (2014) Checklist of helminth parasites of amphibians from South America. Zootaxa 3843: 1-93. [crossref]