Abstract
The Nankai Trough, the region in which the Philippine sea plate and the Eurasian plate meet, is located off the south coast of Wakayama Prefecture, Japan. IODP drilling core samples were collected there and stored at -80 ° C in the Kochi Core Center. It is generally known that damage occurs to organisms in core samples due to the volume expansion of water during the freezing storage step. In this study, we tried to extract the bacterial flora from long-term stored core samples with an attached potential-controlled electrode. As a result, we have extracted approx. 106 cells/g of living bacteria from each core sample and analyzed the living bacterial flora in the stored deep-sea core.
Keywords
Deep sea, Extremophiles, IODP core sample, Microorganism electrical retrieval, Nankai trough
Introduction
Kochi core center (KCC) is one of four International Ocean Discovery Program (IODP) curation centers located around the world. At IODP curation centers, a lot of core samples are stored in deep freezer rooms and are provided to researchers worldwide. These are important resources for researching novel ecosystem communities, earthquake mechanisms, and methane hydrate deposits. Ideally, researchers hope to use fresh cores immediately after collection on ship. On the other hand, we know a lot of important core samples are frozen at several institute centers and we should make good use of them. In this experiment, we tried to extract the bacterial community from long-term stored core samples using an attached potential-controlled electrode. This result shows that approximately 106 cells/g of living microorganisms remained in samples stored at -80 ° C for 12 years, and that it was possible to extract living microorganism flora from the stored core samples.
Materials and Methods
Deep Sea Core Samples
The deep-sea core samples were collected by the submersible boring ship Chikyu from the Nankai Trough, Japan (Table 1 and Figure 1a, b) [1–5]. Cores 1 to 3 were collected from the Eurasia sea plate, Cores 6 to 7 from the Philippine sea plate, and Cores 4 to 5 from close to the Nankai trough (i.e., the boundary region between both plates) (Figure 1b). Prior to bacterial extraction for this research, these samples were stored at – 80 ° C at IODP Kochi Core Center (Figure 2a, b).
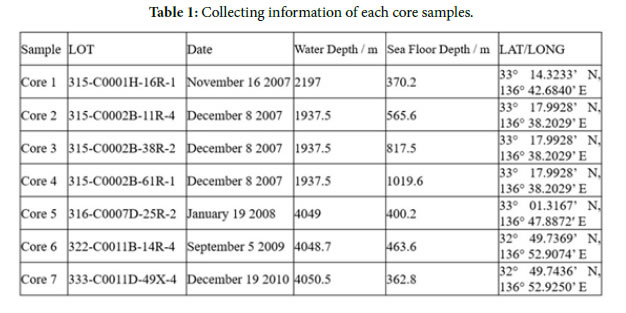
Figure 1. Deep-sea core collected point. a; Boring point by each expedition on the Geographical survey institute map, b; Arrow means core sampling depth.
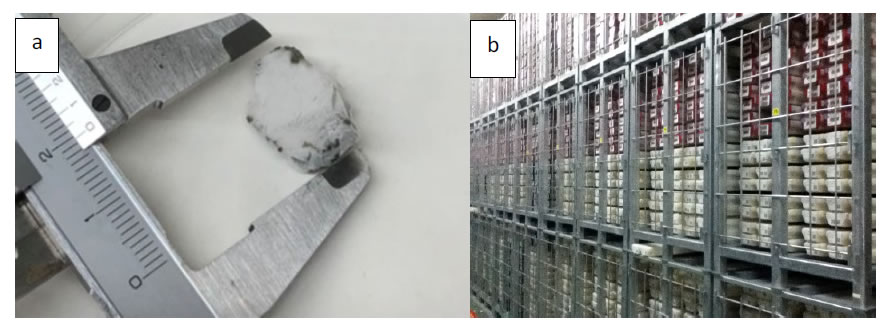
Figure 2. Core Sample and Kochi Core Center (KCC). a; Frozen deep-sea core sample, b; Stored Core Sample in freezing room at KKC.
Ionic Analysis in Deep-Sea Core Samples
The core samples were suspended into PBS(-) to 0.5 g/ml. The ion composition of the deep-sea core was analyzed using the LC-20AP ionic chromatography system (Shimadzu Co.).Negative ions were separated into Shim-pack IC-SA2 (250 mm × 4.0mm) and mobile phase containing 12 mM NaHCO3 and 0.6 mM Na2CO3. Positive ions were separated into Shim-pack IC-C4 (150 mm × 4.6 mm) and mobile phase containing 3 mM oxalic acid as mobile phase. The flows were 1.0 and 1.2 ml/min at 30°C, respectively.
Attachment and Detachment of Uncultured Bacteria in the Frozen Core Sample
Wash of Electrodes and Potential Application
For metagenomic analyses of bacteria in the core samples, living bacteria were collected from the sample using an electrode chamber device. Both the Ag/AgCl electrode and the ITO/glass electrode (ABLE Co., Tokyo, JPN) (Figure 3a) were sonicated in diluted water for 5 min and immersed in 1 M NaOH for 10 min to remove any unwanted deposits. Then, the electrode chambers were sterilized by irradiation with UV light for 5 min in a clean bench.The deep-sea core samples were suspended in PBS(-) to 0.5 g/ml and stored at 4 ° C. To attach bacteria in the core sample to the ITO/glass electrode, 1 mL of the suspended core sample was poured into the chamber system and then topped up with PBS(-) until the solution touched the Ag/AgCl reference and the Pt counter electrode. The constant potential application was done using a potentiostat (ABLE Co., Tokyo, JPN) (Figure 3b). In the deep-sea core sample, −0.4 V vs. Ag/AgCl constant potential was applied to the ITO/glass electrode for 2 h at 8 ° C in PBS(-) [6].
Figure 3. Potential-controlled electrode. a; Ag/AgCl electrode and ITO/glass electrode, b; Potentiostat, c; Bacteria peeling device.
Detachment of Living Extremophiles Flora
The ITO/glass electrode was washed 5 times with PBS(-) at room temperature, and the bacteria attached to the electrode were detached by applying ±10-mV vs. Ag/AgCl 9-MHz triangular wave potential for 60 min with bacteria peeling device (ABLE co., Tokyo, JPN) (Figure 3c) in 10 ml of fresh PBS (-) at 8 ° C.
Fluorescence Microscopic Observation
Microorganisms were observed with a fluorescence microscope in order to count the bacteria population. The staining reagent was prepared using a LIVE/DEADBacLight Bacterial Viability Kit (Thermo Fisher Scientific K.K., Tokyo, JPN) and was added to 1 mL of the detachment sample, and then incubated at room temperature for 15 min. After being trapped on a 0.2 m black membrane filter, the microorganism survival rate was calculated by fluorescence microscopic observation (NIKON Co., Tokyo, JPN) while irradiating with wavelengths of 450 to 490 nm and 510 to 560 nm [7, 8]. Living microorganisms emit green light when irradiated with wavelengths of 450-490 nm due to the Syto9 fluorescence reagent and dead microorganisms emit red light when irradiated with wavelengths of 510-560 nm due to the PI fluorescence probe.
Metagenomic Analysis with NGS
DNA Extraction and Overhang Region Attachment
Genomic DNA of the electrically isolated microorganism in the deep-sea core was extracted using the Cetyltrimethylammonium bromide (CTAB) method [9]. The microorganism 16srRNA v4 region fragments were amplified from the CTAB extracted genomic DNA with universal primers 515F (5’-3’) and 806R (5’-3’) [10] and Q5 High-Fidelity DNA Polymerase following the recommended protocol (New England Biolabs Japan Inc., Tokyo, JPN). Both primers 515F and 806R linked overhang sequence (under line sequence) to the 5’ terminal of primers such as forward primer 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGCCAGCMGCCGCGGTAA-3’ and reverse primer 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHVGGGTWTCTAAT-3’. The following PCR program was used: 98 ° C for 30 sec, followed by 35 cycles of 98 ° C for 7 sec; 57 ° C for 20 s; and 72 ° C for 45 sec, followed by 72 ° C for 2 min. To add the index sequence to the overhang region, each 25 μl aliquot of the reaction mixture contained 5 μl of Nextera XT Index primer 1 and 2 (Illumina, Inc., San Diego, California, US) and 25 μl of 2× KAPA HiFi HotStart Ready Mix (Nippon Genetics Co.,Ltd, Tokyo, JPN) and followed thermal cycler protocol from Illumina co.[11, 12].
Metagenomic Analysis
After purification through AMPure (Beckman Coulter, Inc., Tokyo, JPN), the tagmented DNA, 5 μL freshly 0.2 N NaOH was mixed in microcentrifuge tube in order to denature the DNA.The denatured amplicon DNA was diluted to 570 μL 10 pM using the HT1 and combined with 30 μL 10 nM PhiX library. The prepared samples were then loaded onto the MiSeq, and the MiSeq program was carried out. After MiSeq program was completed, the raw data was analyzed with Geneious software (TOMY DIGITAL BIOLOGY Co., Ltd., Tokyo JPN).
Result and Discussion
Ionic Analysis in Deep-Sea Core Samples
The negative ion SO42- was markedly increased in Core 4 compared to the other core samples. That result might be due to a chemical reaction caused by the high pressure and high temperature conditions resulting from the huge friction at the plate boundary [13, 14]. Moreover, no relationship between sulfate reducing bacteria and SO42- in this Core 4 from metagenomic analysis was found. It is very interesting that none of the results showed a remarkable shift in the microorganism flora such as desulfobacteraceae, desulfobulbaceae and desulfoarculaceae families (Figure 4).
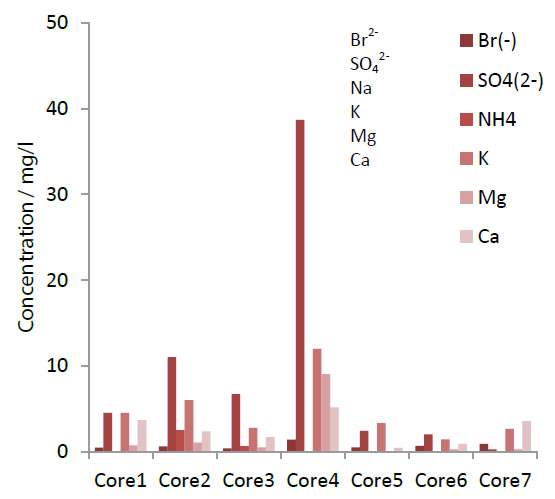
Figure 4. Ion concentration after ionic chromatography.
Living Microorganisms Population and Flora in the Frozen Core Sample
Negative charged electrodes work as electron donors for living microorganisms, so bacterial fimbriae contact with the ITO/glass electrode surface when a negative potential is applied [6, 15-17]. After detachment from the potential-controlled electrode, the average of living bacteria in each core sample was found to be 9.8 × 105 cells/g (Figure 5) and confirmed good agreement with sequence count by metagenomic analysis. In addition, dead cells were also found in amounts corresponding to about 10% of the living cells population. Moreover, the lowest population showed in Core 4, from the deepest seafloor depth (1019.6 m), was probably caused by the extreme environment found there. This result is supported by the metagenomic analysis OTU data (Figure 6).
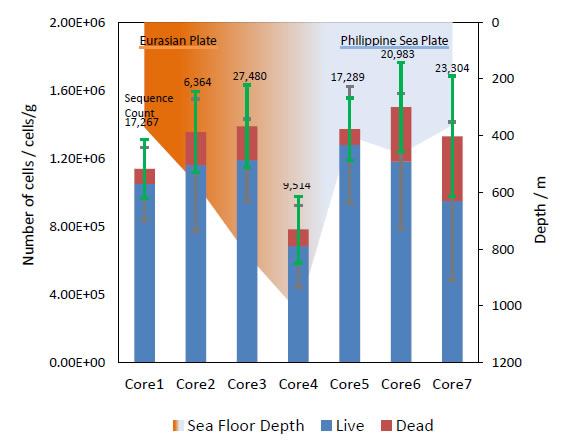
Figure 5. Bacteria population and sequence counts in each core samples after electrical retrieve.
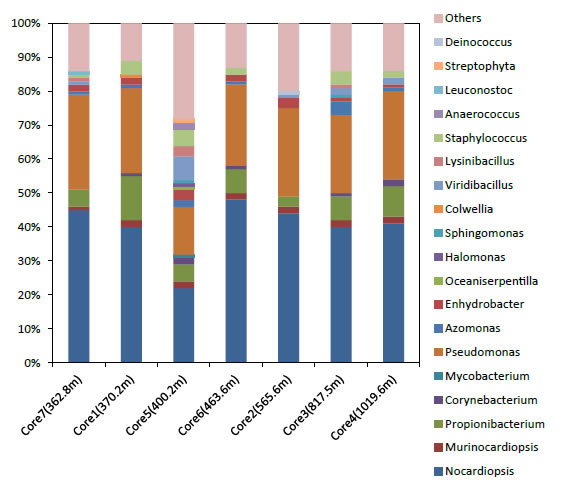
Figure 6. Living bacterial flora after electrical retrieve from each core samples.
We can see the metagenomic analysis results in Figure 5. The flora composition found in Core 5 was remarkably different to the other core samples. It means an increase in the bacterial diversity. This can be seen in the pink “Others” section of the bar graph, which is much larger for Core 5 than in other core samples. Also, other bacteria grew up in this core, such as 0.9% mycobacterium, 1% oceaniserpentilla,1% halomonas, 2% Araerococcusand 1% streptophyta. Core 5 was collected from the shallow boundary region of the both plates, an environment in which it is easy to build up special bacterial flora due to the effect of external factors such as meso-pressure and the two different adjacent plates [18]. Table 2 shows the link of analyzed data by Geneious in this study. It is interesting that the chemical composition and microorganisms flora at the boundary plate region are similar to those found in extreme hot-spots in deep-sea cores. This experiment shows that we can rescue microorganism flora from core samples stored long-term at -80 ° C. The potential-controlled electrode extraction technique shows promise for the development of extremophiles research.
Acknowledgment
The authors would like to acknowledge Kochi Kore Center for supplying deep-sea core samples from IODP Expeditions 315, 316, 322, and 333 around the Nankai trough. We are grateful to Sumihiro Koyama of ABLE Co. and Biott Co. for providing method of bacterial attach and detaching technique. And we thankful for the support given by our laboratory members and David Marsh. This research was supported by KOSEN 4.0 INITIATIVE of National Institute of Technology.
References
- Masataka K, Harold T, Juichiro A, Gaku K, et al. (2009) Expedition 315 Site C0001.Proceedings of the Integrated Ocean Drilling Program.
- Masataka K, Harold T, Juichiro A, Gaku K, et al. (2009) Expedition 315 Site C0002.Proceedings of the Integrated Ocean Drilling Program.
- Masataka K, Harold T, Juichiro A, Gaku K, et al. (2009) Expedition 316 Site C0007.Proceedings of the Integrated Ocean Drilling Program.
- Saneatsu S, Michael BU, Yu’suke K, et al. (2010) Site C0011.Proceedings of the Integrated Ocean Drilling Program.
- Pierre H, Toshiya K, Moe Kyaw Thu, et al. (2012)SiteC0011.Proceedings of the Integrated Ocean Drilling Program.
- Sumihiro K, Masa-aki K, Yukari O, Tetsuya M, Yuji Hatada,et al. (2013) Attachment and Detachment of Living Microorganisms Using a Potential Controlled Electrode.Marine Biotechnology15 : 461-475. [crossref]
- Christine J. Bunthof, Saskia van Schalkwijk, Wilco Meijer, Tjakko Abee,HugenholtzJ, et al. (2001)Fluorescent method for monitoring cheese starter permeabilization and lysis. Appl Environ Microbiol67 : 4264-4271. [crossref]
- Takehiro O, Junji T, Khairul M (2012) Potentials of mouthwashes in disinfecting cariogenic bacteria and biofilms leading to inhibition of caries. Open Dent J 6 : 23-30. [crossref]
- Aboul-Ftooh AMN, Abdel-Sadek OH (2019) Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bulletin of the National Research Centre 43.
- J. Gregory Caporaso, Christian L. Lauber, William A. Walters, et al. (2011)Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample.Proceedings of the National Academy of Sciences of the United States of America108 : 4516-4522. [crossref]
- Bert DR, Milad A, Alice SB, Jos RW,Margot E. Smit, et al. (2014) Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science345.
- Xin Duan, Arjun Krishnaswamy, Irina Huerta DL, Joshua R S (2014) Type II Cadherins Guide Assembly of a Direction-Selective Retinal Circuit. Cell 158 : 793-807.
- Sachiko W, Miho T, Masaru M (2003)Identification and determination of sulfur compounds dissolved in water from volcanic ashes of Mt. Oyama in Miyake island and their effect to the environment BUNSEKI KAGAKU52 : 997-1003.
- Tatsuhiko K (2015) Chemical Composition of Mantle Wedge Fluids.Journal of Geography(Chigaku Zasshi)124 : 473-501.
- Souichiro K (2014) Extracellular Electron Transfer: Microbial Respiration on Solid Compounds. Japanese Society of Microbial Ecology.
- Kengo I (2011) Microbial Extracellular Electron Transfer.Journal of Environmental Biotechnology.
- Gemma R, Kevin DM, Teena M, Julie SN,Mark TT, et al. (2005) Extracellular electron transfer via microbial nanowires. nature435 : 1098-1101.
- Takuro N, Yoshihiro T, Miho H, Shigeru S, Akiko M, et al. (2015) Hadal biosphere: Insight into the microbial ecosystem in the deepest ocean on Earth.Proceedings of the National. Academy of Sciences of the United States of America112.