DOI: 10.31038/JMG.2018112
Summary
Gene transcripts (human FMR1, chicken ovomucoid, human 25 vitamin D3 1-α-hydroxylase, hamster adenine phosphoribosyl transferase and human insulin) were scanned for ESE (Exonic Splicing Enhancer) by ESEfinder 3.0, CSHL. The ESE distribution is gene specific with a consensus pattern.
- The ESEs are more abundant in smaller genes.
- Some of the consensus pattern includes (a) SF2/ASF is clustered in the 1st exon in comparison with the last exon and, SRp55 and SRp40 are more in the last exon in comparison with 1st exon; (b) SC35 and SRp40 are in the central region of the gene body but SC35 is shifted toward the 5’ half of the transcript and SRp40 is shifted more toward the 3’ side; (c) The highest cluster of SF2/ASF (9/10) and SC35 (4/5) are in 5’ side of the molecule and the highest cluster of SRp40 (3/5) is in 3’ side.
- A consensus Alu sequence contains 15–17 ESEs (SF2 + SC35 + SRp40 + SRp55) and is enriched with SF2 with some variations in individual Alu elements.
- Satellite DNAs have more SF2 than other ESEs.
- DNA breakpoints and MALAT1 have more SRp40.
- Repeat sequences have specific ESE enrichment; SF2 in CGG repeats, SRp55 in CAG repeats, SC35 and SRp55 in CUG repeats and SF2 in CCUG repeats, suggesting that these repeat sequences may sequester around certain SR proteins leading to alteration in splicing and gene expression patterns.
1. Introduction
In the post genomic era after completion of the 3 × 109 bp haploid human genome sequence, it is now possible to put it into perspective and assign gene function. With the breakthrough findings of discontinuous genes and splicing in eukaryotes, a mountainous number of factors have been reported to influence the splicing reactions. As the transcription, splicing and transport are intimately interconnected, overlapping functions of some of the factors are inevitable. In addition, mRNA expression in gene constituents ranges from 10 to 15% (hnRNA) and furthermore, lncRNA genes have been identified as often as coding genes or possibly more. The repeat genes comprise ~45% and remaining genes include satellite DNA, cetromeric DNA, telomeric DNA, rRNA genes, snRNA genes, tRNA genes, miRNA genes, 5SRNA genes and other housekeeping genes (Table 1). There are regional differences in the genes in these categories which are difficult to be delineated by base composition or nucleotide sequence. Oligonucleotide characteristics may differentiate some of the regional differences which may confer different functions. The ESEs (Exonic Splicing Enhancers) which were originally found to have roles in splicing and alternative splicing are further implicated in many other functions. These extra functions include (1) SF2/ASF and SC35 in alternative promoter selection and poly (A) site selection [1], and prevention of mutagenic R-loop formation [2], (2) SF2/ASF in translation and mRNA stability, (3) the SC35 in transcription elongation [3], and (4) SRp20 and 9G8 in the nuclear export of mRNA. The SF2/ASF binds to a purine rich element of chicken PKCI-r mRNA, induces instability and reduces its accumulation in the cell [4]. The SF2/ASF, SRp20 and 9G8 have been shown to shuttle between the cell nucleus and cytoplasm. This shuttling function requires RRM and SR motifs to be present in the molecule [5]. Moreover, SF2/ASF enhances translation of mRNA and the presence of ESE element in the mRNA enhances further more [6]. Those shuttling SR proteins (SF2/ASF, SRp20 and 9G8) also function as adaptor proteins for TAP/NXF1 mRNA export [7]. The splicing is carried out in spliceosomes, supra- spliceosomes containing pre-mRNA 5’ and 3’ splice sites, branch sites, enhancer sites, five snRNPs and ~150–300 protein factors mounting to MW 100 million Da. These include hnRNP proteins, snRNP proteins (U1 RNP, U2 RNP, U6 RNP, U4/U5 RNP), DEAD box helicase/ATPase (p68/prp28 etc), enhancer proteins (SF2/ASF, SC35, SRp40, SRp55, 9G8, SRp20, SRp75 etc.) and other proteins. The protein components are different depending on the sequence elements present at the splice site, cell type, developmental stages and pathologic conditions. The splicing codes can be divided into the following two groups: (1) consensus codes and (2) specific codes. In general, the consensus codes are applied to most splicing reactions which include a GU 5’ splice site, AG 3’ splice site, branch site, or five snRNP components. The specific codes may include ESE, ESS, ISE and ISS which regulate splicing patterns involving exon skipping/inclusion as well as defining pseudoexons/non-coding exons. The ESE components and their distributions are species specific and are particularly marked for ISE sequences and distribution in gene transcripts. It is interesting to see that some of the splicing mechanisms in fish transcripts are not applicable in mammalian cells [8]. The tissue specificity of ESEs are demonstrated in SRp55 in calcitonin/CGRP alternative splicing [9], SRp40 in PPARγ1/PPARγ2 alternative splicing [10] in adipocyte differentiation and others. It has been reported that exons contain more ESEs and introns contain more ISS [11]. The SF2/ASF was found to associate with U1–70K protein at the 5’ splice site. The last intron splice acceptor has been shown to influence on transcription termination by exon definition mechanism [12]. The SF2/ASF and 9G8 has been found to enhance splicing of the fibronecting ED1 exon inclusion in a promoter dependent manner [13]. The ESE functions are combinatorial and position dependent [14]. It is therefore of interest to find out whether splicing codes are distributed evenly throughout the genes or there are transitions in splicing code in gene structure as it is transcribed from 5’ initiation site to the elongation and 3’ termination. It has been reported, by chromatin cross-linking and immunoprecipitation (Chip) methods, that SR proteins bind to RNAs and SC35 not only binds to RNA but also crosslinks to DNA as well. The SR proteins are recruited from the pool during transcription and RRM plays an important role in nascent RNA binding. It was demonstrated that SRp20 binding was ~2X more in exon 1 compared to exon 4 and SRp55 binding was ~2X more in exon 4 than in exon 1 of the fos gene transcript [15]. The importance of ESE/ESS and ISE/ISS has been emphasized in pseudogene suppression and regulation of strong and weak exons [16]. Many factors influencing splicing reactions are splice site strength, presence and absence of ESE/ESS, ISE/ISS, regional RNA secondary structure, DEAD box RNA helicases/ATPases, presence and absence of Alu, LINE, PTC (premature termination codon), triplet repeats, dinucleotide repeats and others. In addition, transcription factors acting on initiation and elongation can impact splice site selection. The rate of elongation by RNA polymerase II has been shown to influence alternative splicing events [17]. In view of the fact, that diseases causing DNA mutations may be involved in alternative splicing in 50–62% of the cases [18,19], the importance of studying the regulations of splicing mechanisms cannot be overstated. Moreover, the characteristics of DNA breakpoints are not well defined and more rigorous analyses are needed for the understanding of cancer and other gene- based diseases.
Table 1. Genomic Constituents (3×109bp)
|
Coding Genes (Protein) |
Exons+Introns |
1-2% coding |
Introns; 9-13% snoRNA miRNA |
10-15% of genome |
|
Noncoding genes: 22,000 Pseudogenes; 13,000 |
Exons+Introns |
|
snoRNA miRNA |
Σ~15-25% |
|
Repeat genes (Retrotransposons; Alu, SVA, LINES, LTR, Dinucleotides repeats etc) |
Alu: ~300 bp L1: ~6kbp SVA; 0.7-4 kbp |
~106 copies (~10-15%) ~8.3% (~7-17% ~0.2% |
|
Σ~45% |
|
Satellite DNA(α, β, γ and III) |
|
~1-5% |
|
|
|
rRNA gene |
rRNA Gene 5SrRNA |
~350-500 genes (~15Mbp) |
|
1-2% ~1-5% |
|
SnRNA gene |
U1,U2,U3,U4, U5,U6,U7,U8, etc |
|
|
~1% |
Many essential factors and regulators of gene expression exist at different levels of gene expression. At the level of splicing, a variety of ESE, ESS, ISE and ISS have been reported. These motifs not only function at the canonical splicing but also during alternative splicing, increasing the repertoire of the protein populations. These motifs exist in isolation or in clusters in many different combinations. Accordingly the presence of these motifs and binding proteins can regulate gene expression in a context dependent manner. The SR-proteins which binds to ESEs are found to stimulate splicing reaction in vitro.
In addition to ESE (Exonic Splicing Enhancer), ESS (Exonc Splicing Silencer), ISE (Intronic Splicing Enhacer) and ISS (Intronic Splicing Silencer) were found to enhance or silence splicing reactions in cell extracts with model splicing components as well as in the cells with knock in and knock out experiments. The SR-proteins bind to specific sequence elements in order to exert their functions. Some of the ESE consensus sequences included in this study are 1) SF2/ASF (SRSASGA), 2) SC35 (GRYYcSYR), 3) SRp40 (ACDGS), and 4) SRp55 (USCGKM); where P=Purines (A or G), Y=Pyrimidines (C or U), D=A, G or U, S=G or C, K=U or G and M=A or C [20,21,22]. The distribution of these motifs is essential for specific gene expression during developmental stages and in specific cell types.
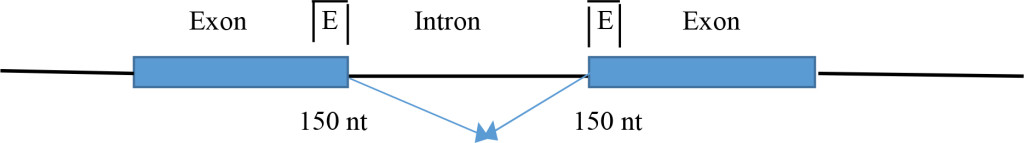
- Distribution in canonical splicing revealed the clustering of ESE at the splice sites for the correct splice site selection and enhancement [1]. The ESEs are clustered within 150 nt from the splice sites in exons.
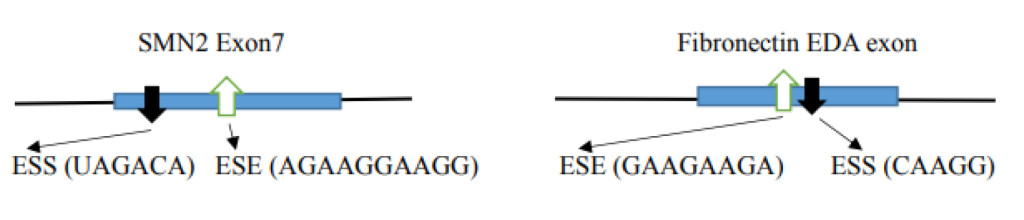
- Distribution of a silencer at the canonical splice site leads to alternative splicing. The example illustrates the SMN 1 gene where variation occurs in one nucleotide (C→U) in the SMN2 gene exon 7 creating a silencer motif (UAGACA; in SMN1 the sequence is CAGACA) and inhibiting a splice to exclude exon 7 in the final product [23]. Another example is in the case of the Fibronectin EDA exon where the presence of ESS excludes the EDA exon and the presence of ESE includes the EDA exon [24].
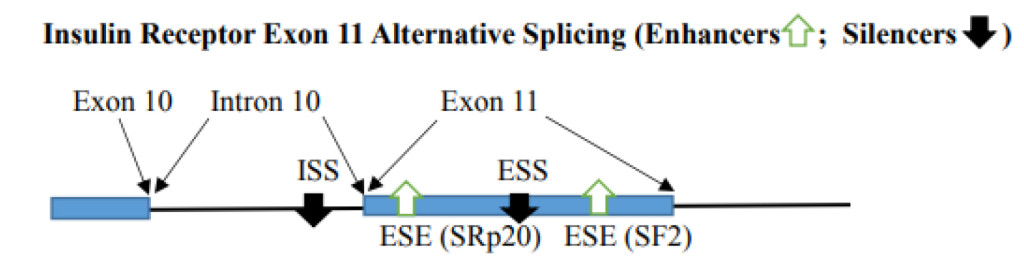
- Variation in Regulator numbers in exon and intron can impact alternative splicing. When inhibitory factors are multimers, the exon is excluded. With one high score ESE, the splicing at the correct site can be enhanced. This depends on the balance of enhancers and silencers. With the availability of binding SR proteins, the splicing can be altered to provide alternative splicing. For example, the Insulin receptor gene contains 2 ESEs [SRp20 (CUCUUCA) and SF2 (CGAGGA)] and ESS (CUGGUGCCG) in exon 11 and additional ISS (CCUCCAAGUGUC) in intron 10. ESE binds SF2 and SRp20 to enhance exon 11 inclusion. The silencers bind CUG-BP1 and cause exon 11 exclusion [25]. Mutation of any one of the ESEs will compromise exon 11 inclusion and the mutation of ESS or ISS result in causing the exon 11 inclusion.
- Position dependent Regulators (ESEs) in non-splicing conditions such as in intron-less mRNA and non-coding RNA are of interest because they impose ESEs in extra functions other than in splicing. The specific ESE motifs are differentially distributed.
It has been widely observed that the extra activities include mRNA transport, localization, translation and metabolism by NMD (nonsense mediated decay). It is of interest to see whether or not different proteins function differently in other splicing reactions. Involved proteins in first and second step splicing reactions are different, and mRNA modifications and processing are different from 5’ initiation of transcription till 3’ end poly (A) formation. The different SR-protein binding sites are differentially distributed in the gene from the 5’ end to the 3’ end. The SF2/ASF is more clustered at the 5’ side of the gene whereas SRp55 tends to be clustered on 3’ side of the gene.
The abundance of ESE is inversely correlated with the size of the gene. The repeat elements have clustering of specific ESEs and clustering of ESEs in Alu elements in the FMR1 gene is observed.
2. Materials and Methods
Transcript sequences were obtained from NCBI and Ensemble release. The human insulin gene (NCBI; J00265), hamster APRT (adenine phosphoribosyltransferase) gene (NCBI; X03603), human 25-hydroxyvitamin D3 1-α-hydroxylase gene (NCBI; AB006987), chicken ovomucoid gene (Ensemble release 43, http://www.ensemble.org), and FMR1 (NCBI; L29074.1) gene sequences were downloaded from the Website. The analyses are made from transcription start sites to the poly A sites and beyond. When there are multiple TSSs, the farthest upstream TSS is included. In human insulin gene (NCBI; J00265) transcript, total 1,430 nucleotides are analyzed which is from the nucleotide 2,186 (TSS) to the nucleotide 3,615 including 465 nucleotides exons and 965 nucleotides introns.
In the hamster adenine phosphoribosyltransferase (APRT) gene (NCBI; X03603) transcript, total 2,251 nucleotides were analyzed which is from nucleotide 330 (TSS) to 2,580 including 881 nucleotide exons and 1,370 nucleotide introns.
In the 25 hydroxyvitamin D3 α-1-hydroxylase (25VD3H) gene (NCBI; AB006987) transcript, a total of 4,825 nucleotides were analyzed which is from nucleotide 285 (TSS) to nucleotide 5,109 including 2,551 nucleotide exons and 2,274 nucleotide introns.
In the ovomucoid gene transcript, a total of 6,067 nucleotides were analyzed which includes 1,424 nucleotide exons and 4,643 nucleotide introns. The sequence is from TSSII which is 85 nucleotides upstream from the major transcription start site (TSSI). The TSSI is 53 nucleotides upstream from the AUG translation initiation site. The ensemble release is 5,587 from ATG to the end of the 1st poly(A) site and beyond. Adding 342 nucleotides which contains 2nd poly(A) site and beyond (Gerlinger et al., 1982) adds up to 6,067 nucleotides (85 + 53 + 5,587 + 342 = 6,067). This sequence includes a 138 nucleotides 5’UTR, 633 nucleotide exons, 4,643 nucleotide introns and a 653 nucleotide 3’UTR.
In the FMR1 gene (NCBI; L29074.1) transcript, a total of 39,224 nucleotides were analyzed which is from nucleotide 13,652 (TSSIII) to nucleotide 52,875, including 4,456 nucleotide exons and 34,768 nucleotide introns.
ESEs have been screened by ESE finder 3, Cold Spring Harbor Laboratory [22] with a default threshold, otherwise stated. The default thresholds scores are SF2/ASF; 1.956, SF2/ASF (IgM-BRCA1); 1.867, SC35; 2.383, SRp40; 2.67 and SRp55; 2.676. The number of ESEs were analyzed in the total transcript, exon only, intron only and splice sites. The number of ESEs are expressed as the number of ESEs per 100 nucleotides in each categories. The number of ESEs in noncoding RNAs were analyzed accordingly. The noncoding RNAs analyzed are:
- The DNA breakpoint sequences are from the reference by Lui et al., 2011 [26].
- The NEAT 1 sequence is from NCBI Reference sequence NR_028272.1
- The MALAT 1 sequence is from NCBI Reference sequence NR_002819.3
- The α-satellite consensus sequence 1 and 2 are from reference by Waye and Willard [27] and consensus sequence 3, 4 and 5 are from Vissel and Choo [28].
- The Alphoid sequence (334 bp) is from GenBank S49988.1.
- The β-satellite sequence (955 bp) is from GenBank M81228.1.
- The γ-satellite DNA (1962 bp) is from GenBank X68546.1.
- The satellite III in chromosome 14 (1,404 bp) is from GenBank S90110.1.
- Alu Major, Alu Precise and AluPV (HS) from reference by Maraia et al., 1993 [29]
- Human Y RNAs from reference by Christov et al., 2006 [30]
- Sleeping beauty sequence from Hackett et al., 2004, von Pouderoyen et al., 1997 [31, 32].
- ESE clustering in short repeat sequences such as CAG, CUG, CCUG and CGG have been analyzed [33]
3. Results
3.1 ESE Distribution in Coding Gene Transcript
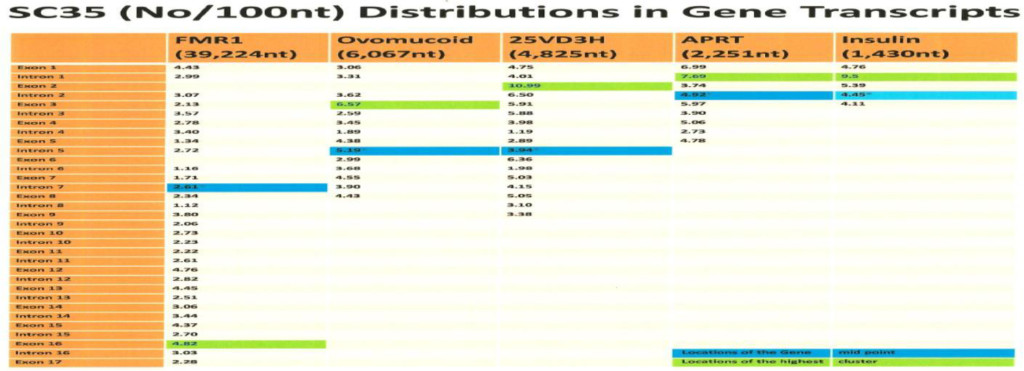
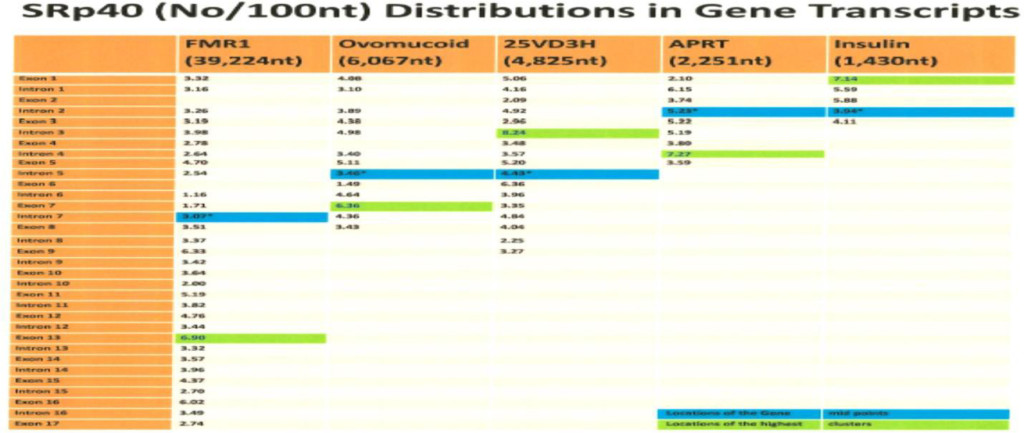
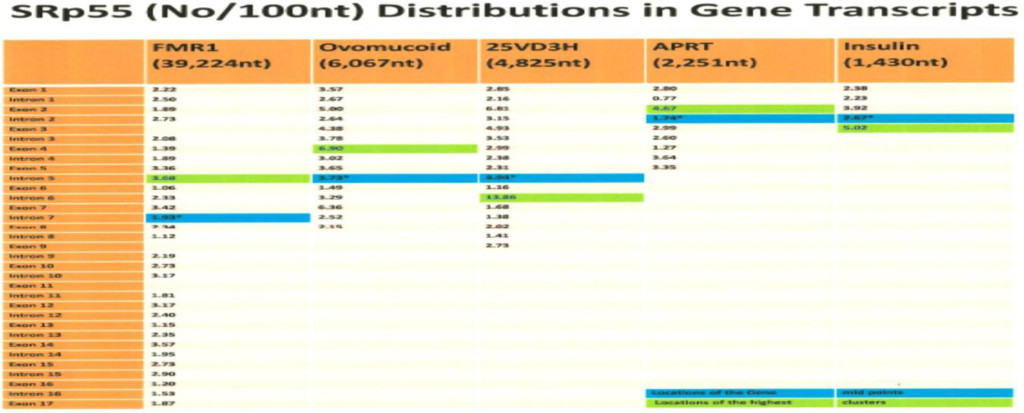
The computer screening of ESE distribution in pre-mRNAs revealed gene specific distributions as well as some consensus patterns of distribution. Using ESEfinder 3 (CSHL), the distribution of SF2/ASF, SC35, SRp40 and SRp55 have been screened. An example of ESE distribution is shown in Fig. 1. Table 2a, Table 2b and Table 2c summarize the ESE distributions in five different gene transcripts. The total number of ESEs (SF2/ASF+SC35+Srp40+SRp55) in each gene showed an inverse correlation with the chain length of the gene transcript, where the shortest gene transcript of insulin gene has more ESEs than the longer gene transcript of FMR1 per 100 nucleotide bins (Table 2a). The clustering of ESEs at splice sites is evident in short gene transcript. However, in longer gene transcripts, like the FMR1 gene, no differences are found between splice sites and other regions in gene transcript (Table 2a). In overall counts [total length 53,797 nucleotides (FMR1+Ovo+25VD3H+APRT+Insulin)], SC35 and SRp40 are more abundant and SRp55 is the least abundant in 5 gene transcripts examined (Table 2b). Individual gene transcript have their characteristic content of ESEs where 25VD3H and insulin gene transcripts have SF2/ASF abundance (Table 2c). These ESEs are clustered at certain regions of the gene transcripts, for example the first exon has more abundance of SF2/ASF (Table 3a) whereas the last exon has relatively more SRp55 in comparison with other ESE motifs (Fig. 2, Table 3b). Overall, the ESEs are more abundant in first exons of all 5 genes tested (Table 3a) and ESEs are less abundant in last exons (Table 3b). The SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing [9].
Although there is focal regional clustering, 5’half and 3’half analyses do not reveal significant differences. Analysis of ESE distribution throughout the genes, exon by exon and intron by intron revealed the more clusterings of SF2/ASF at the 5’ region, SC35 in the 5’ side central region while the SRp40 was in the 3’ side central region and SRp55 in the 3’ region of the gene transcripts. Examples are illustrated in (Figures 3 a,b,c,d)
The role of ESEs is critical for the formation of specific gene products. The changes in gene structure by point mutations (SNP) or indel (insertion/deletion) leads to changes in ESE distribution which produces variant mRNA products. The insulin gene at chromosome 11 contains three exons and two introns (Figure 1). The translation initiation site is in the exon 2 which leaves exon 1 and part of exon 2 as a 5’ UTR. Even in normal pancreas β-cells, it was found that ~10% of the insulin mRNA contains extra 26 nucleotides in the 5’ UTR which is derived from alternative splicing at cryptic 5’ splice site at position 68 in intron 1 when compared with canonical splicing at position 42 (Figure 1). The proportion of the longer 5’UTR containing insulin mRNA increases markedly in prolonged hyperglycemic condition and has a higher efficiency of translational activity [34]. In African population, the variant gene with TTGC insertion at the position close to 5’ splice site of intron 1 (47–50) leads to attenuation of canonical splice site at the position 42, higher proportion of the mRNA is spliced at the cryptic site at position 72 (position 68+4 nt insertion=72) (Figure 4), and has a longer 5’UTR which is 30 nucleotides longer than normal insulin mRNA. The UUGC insertion also changes ESEs distribution, where extra SRp40 and SRp55 are created (Figure 5) which may also contribute to enhanced translation of a longer insulin mRNA [35, 36]. The activity appears to be more specific to SRp40 and SRp55, because other SR proteins such as SF2 have some promoting activity, but SRp40 and SRp55 have a significantly higher proportion of translation promoting activity with longer insulin mRNA.
It is known that SRp40 and SRp55 have promoting activity on HIV1 genomic translation [37]. Although the exact mechanism of action is not known, the presence of RRE (Rev Response Element) or CTE (constitutive transport element) in viral RNA and specific coding sequences is required for the enhanced translation. Moreover, the RRM2 motif and SR domain in SR proteins are required for the activity. RRM is an RNA binding motif and the SR domain interacts with other proteins, or also with other nucleic acids. The abundance of SR binding sites are present in RRE and CTE. The SF2 sites are the most abundant but a considerably higher proportion of SRp40 sites are also present (Table 4). The SRp40 and SRp55 also increase the proportion of un-spliced RNA for an in vitro splicing condition [37]. In the case of fibronectin (FN) EDA exon (one tissue specific alternative exon of FN mRNA; It is selectively excluded in hepatocytes and included in various extents in other cell types) containing a mRNA construct, the SF2/ASF is the most translation promoting SR protein and the enhancing translation is by increased mRNA utilization by polysomes, translation machinery. The mRNA in translating ribosomes is increased as well as SF2/ASF’s association with translating ribosomes [6].
3.1.1 Characteristics of Individual Gene Transcript
3.1.1.1 FMR1 Gene Transcript
The FMR1 gene has different gene structures from others in that it contains triplet (GGC ) repeats, eight Alu elements, one LINE sequence and four potential microRNA sequences. It has specific characteristics of ESE distribution.
- The triplet repeats in FMR1 contain clustered SF2/ASF (from nucleotide position 100–300) of 36 SF2/ASF motifs (18/100 nt).
- In intron 2, the nucleotides from 12,599 to 12,641 (43 nt) contain GU and AU rich sequences and a cluster of 19 SRp55 (44/100 nt).
- Alu and LINE elements which are jumping genes have more ESEs than exons in the FMR1 gene transcript (Table 5). Although there are consensus patterns such as higher abundance of SF2/ASF than other ESEs, in general, equal numbers of ESEs in total which is ~16 ESEs per 100 nucleotides. The Alu elements have been reported to be enriched in the gene-dense-chromosomes such as chromosome 19 and also more abundant in euchromatin areas than in heterochromatin areas [38]. In view of the fact that there are many exonizations of Alu and LINE elements during evolution [39, 40] it is interesting to observe the presence of high densities of ESEs in addition to 5’(+ oriented Alu) and 3’ (- oriented Alu) splice sites in these elements.
The involvement of Alu and LINE elements in chromosomal inversion have been reported [41]. - The ESE densities in the FMR1 gene transcript are in the order of Alu (15.69/100 nt) > LINE (14.22/100 nt) > Exons (10.97/100 nt) > Introns (10.47/100 nt) > Splice sites (9.87/100 nt).
- It is interesting to observe that where there is alternative splicing in the FMR1 transcript, there are more ESEs; exon 10 has 12.73 ESEs/100 nt., exon 15 has 20.77 ESEs/100 nt. and exon 17 up to stop codon has 16.26 ESEs/100 nt in comparison with 10.53 ESEs/100 nt in the total sequence (39,224 nt) of FMR1. The whole exon 17 is 2,409 nucleotides and up to the stop codon is 160 nucleotides.
The FMR1 gene has more alternative spliced products among five genes studied. The alternative splicing is mostly located at the 3’ half of the molecule involved in exons 9, 10, 12, 14, 15 and 17 [42, 43]. Of these, exon 15 has three 3’ splice sites (Fig. 6 and 7). The ESEs appear to be clustered more around the exon 15 (Figure 6) and 3’ splice site strength are correlated with the amount of spliced products formed. The canonical splice product is the most abundant, and alternative splice site 2 and 3 usages is much less [43-46] (Figure 7). Although there are high strength 3’ splice sites in close proximity to alternative splice site 3, it is not operative in a splicing reaction. It is interesting to observe that there are more silencer motifs present in this region (Figure 8) which may counter-act the splice site operation.
3.1.1.2 Ovomucoid Gene Transcript:
The number of ESEs/100 nt (SF2/ASF + SC35 + SRp40 + SRp55) at splice sites (100 nt at GU containing region + 100 nt at AG containing region) are in the order of splice site 5 (20.5) > splice site 6 (18.0) > splice site 7 (17.3) > splice site 3 (13.0) > splice site 1 (12.5) > splice site 4 (12.0) > splice site 2 (11.0) which are consistent with the fact that intron 5 and 6 are removed earlier than other introns (Table 6). However the order of intron removal of the rest of the introns are not in accordance with the experimental order of intron removal of 5/6 > 7/4 > 2/1 > 3 [47]. The facts may indicate involvement of some other factors in splicing mechanisms such as thermodynamics of secondary structures, RNP stabilities and others [48].
3.1.1.3 25VD3H, APRT and Insulin Gene Transcripts
These genes are not typical hnRNA type pre-mRNA which is comprised of only 10–15% of coding region. Instead the 25VD3H has 52.9% exons, the APRT has 39.1% exons and the insulin gene has 32.5% of exons (Table 2a). In these genes, the shorter the gene the more ESEs content was observed. In addition, ESE densities are higher at the splice sites which were not observed in FMR1 and the ovomucoid genes. It is interesting that the average ESE of 5 genes are more abundant at 5’ splice sites than 3’ splice sites (Table 7) suggesting that the 5’ splice sites are the driving force for spliceosome formation.
In summary ESE screening revealed the following facts:
- The ESEs are more abundant in smaller gene products than large genes and more abundant in the order of splice sites (16.99/100nt)>exons (16.37/100 nt)> introns (15.86/100nt).
- The ESE distributions in transcription unit are gene specific with some consensus such as
- SF2/ASF is clustered more in 1st exon (prevents R-loop formation),
- SC35 is more clustered in 5’ half and the central region (elongation and maintenance of transcription by recruitment of pTEFb). The SC35 has active role in transcriptional elongation [3].
- SRp40 and SRp55 are more clustered in the central and 3’ half regions.
- Mutations including SNP and indel (insertion/deletion) change ESE distribution and produce variant mRNAs
- ESEs and 5’/3’ splice sites strength influence alternative splicing.
- ESS are also abundant in the genes which may counter-act to ESE and valance of ESE/ESS may become operational for successful splicing.
3.1.2 ESE Distribution in Non-Coding RNA Transcript
Abundant SR protein binding sites regulating splicing reactions are found not only in intron containing transcripts, but also found in non-intron containing pre-mRNAs as well as noncoding RNAs. Accordingly, it was found that ESEs have extra functions other than in splicing.
The multi-exonic and mono-exonic intergenic lncRNAs (long non-coding RNA) are identified in human which numbered 14,484 multi-exonic and 46,517 mono-exonic sites [49]. Although the intronic lncRNAs have less sequence conservation, the clustering of ESEs at the splice sites appear to be mirrored the protein coding genes. The exons have high GC content.
3.1.2.1 Repeat DNA Sequence Elements
This category of DNA is comprised of ~45% of the total genome and most of them are silent and a few of them are transcribed in normal condition. The changes in their expression (either increased or decreased) are observed under cellular stress conditions such as after heat shock treatment.
Of these genes, Alu (~10–15%) and LINE1 (~7-!7%) are predominant elements. The Alu repeat is composed of 281 nucleotides with the components of left half and right half. The LINE1 repeat is ~5–7 kb, mostly truncated and non-transcribed. Only ~30 to 100 copies are active in producing endonuclease and reverse transcriptase for retro-transposition (transposase). Other genes in this group include human YRNAs, SVA, di/tri-nucleotide repeats and others. The Alu RNA and hYRNAs are transcribed by RNA polymerase III and multiple dispersed loci are found to produce scRNA [29]. The hYRNA genes are clustered in chromosome 7 [50, 51]. Increased Alu expression has a multitude of effects on other gene expression. In general, these compartment of DNA sequences have approximately equal numbers of ESEs in total ~15 to 17 (SF2/ASF+SC35+SRp40+Srp55) per 100 nucleotides (Table 8).
(a) ESEs in Alu RNA:
The rat Novikoff hepatoma 4.5S RNAI, the first nuclear small RNA sequenced, is identified as a rodent repeat element of human Alu RNA homolog.
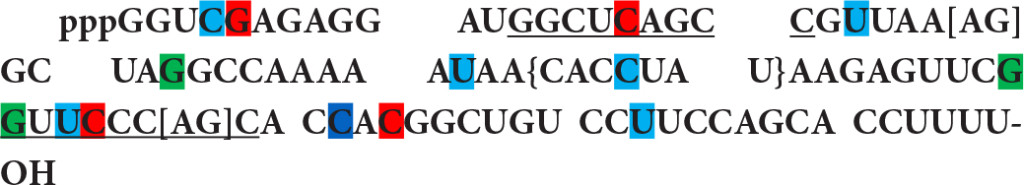
This RNA contains the RNA polymerase III promoter box A and box B like motifs (underlined) and shows interesting enhancer motifs resembling an Alu element transcript. There are 4 motifs of SF2/ASF (first nucleotide is marked in red), 3 motifs of SC35 (green), 6 motifs of SRp40 (bleu) and one motif of SRp55 (navy). It also exhibits 3’ splice sites, [AG] at nucleotide 27 and [AG] at nucleotide 67 as well as 10 branch sites with a highest score +3.15630 at nucleotide 45 {CACCUAU}. In comparison with known Alu elements in FMR1 gene, the resemblance of 4.5S RNA I in ESE, 5’ SS, BS (branch site), and 3’ SS distribution (Table 8) suggests that 4.5S RNA I is more likely an Alu gene expressed in Novikoff hepatoma cells.
Alu class transposons contain ~15–18 total ESEs (SF2+SC35+SRp40+SRp55) with some differences in dominant ESE clustering. Most of them (+ orientation) is dominated by SF2 and 5’ splice sites while Alu (-) has SC35 domination and branch site with 3’ splice sites. Human Y RNA and rat 4.5S RNA I have SRp40 clustering domination. The SRp55 is least abundant in all classes of Alu elements. The cytoplasmic human Y RNAs have been shown to be involved in chromosomal DNA replication [30].
The Alu element has been shown to have many different functions in DNA replication, transcription, splicing (canonical, altenative, exonization and others), gene insertions (transposons) and others [52]. It is interesting to observe that (+) oriented Alu sequences have more 5’ splice sites but the (-) oriented Alu sequences have more 3’ splice sites. It may suggest that exonization may occur from 5’ side of (+) Alu elements and 3’ side exonization from (-) Alu elements. The SRP RNA (7SL RNA) has Alu elements in the molecule [53]. The 7SL SRP (Signal Recognition Particle) is involved in nascent protein guidance into secretory vesicles. In addition, the Alu element in SRP RNA (7SL RNA) is involved in retroviral packaging such as in HIV-1. The retroviruses have been shown to contain host RNA packaged within it, especially the 7SL RNA by interaction with viral Gag protein [54]. It is estimated that there are ~3 to 4 fold molar excess 7SL over monomer of MLV genomic RNA and ~6 to 7 fold molar excess of 7SL over HIV genomic RNA in viruses.
3.1.2.2 ESEs in MALAT1, NEAT1 and DNA Breakpoints
The long non-coding RNAs such as MALAT1 (NEAT2) and NEAT1 are found in nuclear speckles and paraspeckles respectively in the nuclei [55]. The plethora of long non- coding RNAs are known to be synthesized in the enhancer region, intergenic region, intronic region, imprinting region, X-chromosome inactivation region, region of antisense transcription in the gene and others. They have different special functions in enhancer activity, chromatin interactions, transcription, processing/splicing, transport, stability of RNA transcripts, miRNA sequestration, translation and others.
(a) The MALAT1 (Metastasis Associated in Lung Adenocarcinoma Transcript 1) RNA
The MALAT1 RNA is an lncRNA with a chain length of 8,758 nucleotides (NCBI; NR_002819.3) and the gene is located at the chromosome 11q13. The gene is expressed highly in lung, pancreas, prostate, ovary, colon and other normal organs.
The MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1), also known as NEAT2 (Nuclear-Enriched Abundant Transcript 2), is an abundant nuclear RNA. In association with SC35, it is localized in nuclear speckles (IGC; Interchromatin Granule Cluster) [55, 56]. It is expressed in NSCLC (Non-Small Cell Lung Cancer) and the MALAT- 1 expression is increased three-fold in metastatic NSCLC, and in some cases (6 cases from 23 cases) in association with loss or gain of the chromosome 11q region. The high expression of MALAT-1 is associated with a poor prognosis and worse survival [57]. The MALAT- 1 is upregulated in high Gleason score and castration resistant prostate cancer and the siRNA against MALAT-1 inhibited prostate cancer cell growth [58].
The MALAT1 RNA regulates splicing, alternative splicing, nuclear organization, epigenetic regulations; and is known to be involved in human diseases especially in cancer. In lung cancer, MALAT1 actively regulates metastasis associated gene expression and increases cell motility without an effect on splicing. The ASO (antisense oligonucleotide) to MALAT1 prevents metastasis after tumor implantation in mouse xenograft model [59]. The regulation of splicing is involved by sequestration and distribution of SR proteins. The nuclear speckles contain not only SC35 but also SF1, SF2, B”-U2 snRNP, PRP6, SON (SR-related protein) and others. The depletion of MALAT1 increases dephosphrylated form of SR-proteins (inactive) leading to alternative splicing [56]. The MALAT1 gene location at chromosome 11q13.1 is also associated with chromosome breakpoint in renal cell carcinoma [60]. The ESE distribution in MALAT1 has the same pattern as in the DNA break points reported in various genetic diseases [26]. The MALAT1 and DNA breakpoints have the SRp40 clustering domination and SRp55 is the least abundant. They have the smallest number of ESEs in total (Table 9). It is interesting to point out that SRp40 motif sequence is ACDGS (where D = A, G, or U and S = G or C) and one of the sequences can be ACUGG [20,21,22]. The CUGG motif is also present at the PSS (PGBD5-specific signal sequence; CTGGAATGCAGTG). The PGBD5 is a transposase elevated in pediatric solid tumors and responsible for the gene re-arrangement[61]. Low abundance of SF2 clustering is observed in MALAT1 and DNA breakpoints. The SF2 has been shown to prevent mutagenic R-loop formation [2]. The example of ESE screening in MALAT1 transcript is illustrated in Fig. 9. Although the MALAT1 RNA is processed by 3’ end cleavage by RNase P and the end is stabilized by triple helix, the transcript has numerous 5’ SS, BS, 3’ SS as well as poly (A) sites. The ESEs are clustered at 5’ and 3’ splice sites (marked by arrows in Figure 9). Whether these sites are operational or inactive is not known. There have been at least 10 alternatively spliced small isoforms reported.
This group of sequences have at least total ~11/100nt ESEs and SRp40 dominates over others. The less abundance of SF2 and SRp55 are observed in DNA fragile region of the genome.
(b) NEAT1
The NEAT1 RNA is transcribed from chromosome 11q13 region (multiple endocrine neoplasia locus) and overexpressed in many cancer cells. In prostate cancer cells and breast cancer cells, the NEAT1 expression is increased in ERα dependent manner and the increased NEAT1 changes the chromatin architecture at the promoter site, increasing transcription for the cancer progression. Knockdown of NEAT1 leads to inhibition of cancer progression in prostate cancer [62], and inhibition of growth and apoptosis in breast cancer cells [63].
The NEAT1 and MALAT1 are associated with active genes. The NEAT1 is present at both TSS (transcription start site) and TTS (transcription termination site) while the MALAT1 is present at the TTS and gene bodies (West et al., 2014). In the NEAT1, SF2/ASF is dominating while total number of ESEs (SF2+SC35+SRp40+SRp55) is nearly equal to MALAT1 (Table 9).
3.1.2.3 ESEs in Satellite DNA
Satellite DNA Gene Expression
The satellite DNAs are mostly located in the heterochromatin areas such as peri-centromeric area and sub-telomeric area. These include α-satellite, β-satellite, γ-satellite, satellite 1, 2, 3, 4, 5 and others. The α-satellite constitutes ~5% of total human genome and its monomer is ~171 base pairs. The tandemly repeated sequences are present at the heterochromatin and centromere forming kinetochores. A subfamily of α-satellites are present in acrocentric chromosomes 13, 14 and 21 [64]. The human β-satellite DNA, isolated by Waye and Willard [65] showed diverged ~68 base pair monomer repeats with base composition of G+C in a range of 39–51%. They cloned two β-satellites, pB3 and pB4, and characterized them. The pB3 β-satellites are present only in the human chromosome 9 centromeric region and the pB4 β-satellites are present more widely among acrocentric chromosomes 13, 14, 15, 21 and 22 and others. In acrocentric chromosomes, the β-satellites are present both proximal and distal to rRNA gene clusters [65]. The satellite I, II and III are present at the pericentromeric region of human chromosomes 3, 4, 9, 13, 14, 15, 21 and 22 [66]. The gamma satellite DNAs are present at the pericentromeric region of human chromosome 8, X and Y. It is composed of GC- rich 220 bp unit of tandem arrays [67]. The human gamma-satellite DNA arrays contain CTCF and Ikaros binding sites [68].
These compartments of DNAs are also rarely expressed and their marked changes can occur upon stress and other alterations in cellular condition. An example is demonstrated in HeLa cells upon heat shock at 42oC in comparison with the cells at 37oC. Under this condition, the transcription of sense RNA from satellite III increased >10 times above normal while antisense RNA transcription diminished 2-fold. The transcript remained associated at the transcription site at chromosome 9q12 forming stress granule. The transcription is by RNA polymerase II and HSF1 (heat shock transcription factor 1) is responsible for the increased transcription [69]. As detected by FISH (Fluorescent In Situ Hybridization) in transitional cell carcinoma of the urinary bladder, the pericentric satellite at 9q12 is often lost early in cancer progression [70]. The association of chromosomal fragility (chromatid breaks, chromosome break, chromosome arm loss and others) at the band 9q12 and triple A syndrome (alacrima, achalasia and adrenal insufficiency) is observed, although the AAAS gene is identified at the chromosome 12q13 [71].
Gamma Satellite DNA in mouse is transcribed in developmentally regulated manner. In mouse, cassini which belongs to the γ-satellite/major satellite is up-regulated in drug (Vincristine) or heat shock treated ALL (Acute Lymphoblastic Leukemia) cells [72].
Overall, in satellite DNA, the total number of ESEs vary widely in their distribution. However, SF2/ASF is dominating in all of the different satellite DNAs (Table 10). A different class of satellite DNA has a wide range of different numbers of ESE elements suggesting different satellites have different functions. However the consensus is the high incidence of clustering of SF2/ASF in all the satellite DNA. The SRp55 clustering is relatively high in this group of DNA.
3.1.2.4 ESEs in Short Repeat Sequences
The expanded CUG repeats or CAG repeats in untranslated regions of mRNA have profound effects on cellular metabolism by RNA foci formation. In myotonic dystrophy, the MBNL sequestration leads to aberrant splicing of pre-mRNAs [73]. It is interesting to see that CUG repeats have clustering of SC35 and SRp55 ESE elements which maybe involved in SR protein sequestration. The CAG expansion has the same effect as CUG expansion and CAG expansion has SRp55 clustering (Table 11).
The ESE distributions in non-coding RNAs are summarized as follows:
- The consensus Alu sequence contains 15–17 ESEs (SF2 + SC35 + SRp40 + SRp55) per 100 nucleotides. The SF2 dominates over other ESEs but there are variations in individual Alu elements. (Table 8).
- Satellite DNA has SF2 domination over other ESEs (Table 10)
- DNA breakpoints and MALAT 1 have lesser ESEs and SRp40 dominates over other ESEs (Table 9).
- Repeat sequences have specific ESE enrichment: SF2 in CGG repeats, SRp55 in CAG repeats, SC35 and SRp55 in CUG repeats, and SF2 in CCUG repeats (Table 11).
4. Discussion
The surprising finding of the discontinuous gene structure and the necessity of removing of intervening sequences led to the discovery of spliceosomes and their regulatory factors. A group of SR proteins (serine/arginine rich proteins) have been found to have critical impact on the precision of splicing reactions for correct protein production. This group of protein was found to have a role in splice site recognition and enhancement of splicing reactions at the given site. Additional surprising facts are the presence of these elements not only in intron containing pre- mRNA, but also in non-intron containing mRNA, as well as in non-coding RNA transcripts. Accordingly, the functions of SR-proteins were expanded not only in splicing reactions but also in extra functions in addition to splicing. In fact, SR proteins are involved in all the steps of RNA metabolism including, transcription, DNA stability, splicing, maturation, transport, and translation.
When transcription is activated, SR proteins are enriched around the transcription sites [74]. The SR proteins also bind to histone H3 tails in a dynamic manner [75]. They are directly involved in transcription at the initiation, elongation and termination sites. The SF2/ASF has been found to prevent R-loop formation [2, 76] at initiation as well as during transcription, thus leading to protection of DNA from cleavages. The SC 35 has a p-TEFb activation function facilitated by binding to ESE in nascent transcripts, recruitment of p-TEFb-7SK RNP complexs and release of p-TEFb from 7SK RNP [77].
4.1 ESE function in Splicing
The splicing reactions include (a) canonical splicing and (b) alternative splicing.
4.1.1 ESEs in canonical splicing
A large number of splice site analyses by SR protein binding by the CLIP method revealed the ESEs are clustered at the exons within ~150 nucleotides of the splice site [1]. The functions of ESE in constitutive splicing include:
(i) Correct splice site selection and enhancement in constitutive splicing, (ii) Enhancement of weak splice site splicing, and (iii) Enhancement or suppression of splicing in a context dependent manner. The ESE works as an individual SR protein or together with other proteins (SF2, SC35) at the site, and each ESE regulates a different group of splicing events.
The SF2/ASF has been shown to require first-step splicing and bimolecular ligation of 5’ and 3’ splice sites in the initial phase of a second-step splicing reaction. The SF2/ASF requirement is demonstrated in the IgM pre-mRNA M2 exon, Cis-splicing of HIV-tat exons 2 and 3, and β-globin exons 1 and 2 [78]. Different ESEs have differences in their activities in specific pre-mRNAs.
4.1.2 ESE in Alternative Splicing
Alternative splicing is one of the major causes of diversity in protein production from ~25,000 hnRNA coding genes. Errors in alternative splicing is also a dominant causes of diseases. Some of the factors involved in differences in splicing are the ESE (exonic splicing enhancer), ESS (exonic splicing silencer), ISE (intronic splicing enhancer) and ISS (intronic splicing silencer) regulators. The specific SR-proteins have specific alternative splice site selections in specific cells and during developmental stages. The mammalian gene construct with multiple introns confers more than 90 to 95% alternative splicing, producing expanded diversities of protein production. Alternative splicing is regulated by ESEs present in pre-mRNA sequences and single or multiple SR proteins which contain one or two RRMs at their N-terminal region with specific sequence element binding abilities. The knock down of certain ESE binding proteins revealed that ESEs not only enhance the splicing but also inhibit the splicing, and both depend on the specific context. The alternative splicing includes alternative 5’ selection, exon exclusion, intron inclusion, alternative splice site selection in the exon or intron, and alternative poly (A) site selection. One of the well worked out cases of disease, due to a splicing variation, is in SMA (spinal muscular atrophy). Humans have the SMN1 and SMN2 genes at chromosome 5q13.1. The SMN2 has one nucleotide difference at the position +6 in exon 7 from SMN1 where it is U in SMN2 and it is C in SMN1. This difference in SMN2 acts as an ESS (exonic splicing silencer) which causes the exclusion of exon 7 in the final product which is incompatible with a full length SMN1. This exclusion causes a disease SMA when homozygous loss of SMN1 is present [23].
i) Alternative Promoter Selection
These changes in promoter selection were most affected by downregulation of SRp54. Fewer changes occurred by Rbp1L downregulation among the following 8 ESE binding proteins: B52, SRp54, XL6, SF2, SC35, Rsf1, Rbp1L and Rbp1. Examples include the distal promoter usage being reduced in the Nfat gene when XL6 or B52 are reduced, while the proximal promoter usage is increased in the Indy gene when XL6 or B52 are reduced [1].
ii) Alternative Splice Site Selection
The mechanism of ESE effects on alternative splicing depends on the presence of ESE, ESS, ISE and ISS. The splicing stimulatory factor binds to ESE and stimulates correct splicing, while competitive splicing inhibitory factor binding to the same locus, or close proximity to it, leads to exon skipping.
An alternative splicing example occurs in the Fibronectin EDA exon (also called EIIIA or EDI). The alternative splicing is tissue specific in hepatocytes where EDA is always excluded. In other tissues varying proportions of EDA inclusion and exclusion are observed. Using minigene constructs containing a EDA exon in which ESE (GAAGAAGA) and ESS (CAAGG) were included, it was found that in the absence of ESE, EDA is excluded. However, in the absence of ESS, 100% of the transcripts included the EDA exon [24]. Another factor involved in alternative spicing is mRNA modification. The m6A is present most frequently close to the stop codon and ~50 nucleotides upstream from the cleavage site for polyadenylation [79]. The presence of m6A provides the binding site for YTHDC1 [nuclear m6A binding protein with YTH domain (Tyrosine, Threonine, Histidine)] and in collaboration with SRp20 (SRSF3), it enhances inclusion of an m6A containing exon. On the other hand, SRSF10 (SRp38) enhances exclusion of m6A containing exons in splicing. The YTHDC1 binds to SR-proteins of SRp20 and SRp38 but not other SR-proteins. It is dependent on SRp20 and SRp38 binding sites which act in close proximity to m6A in pre-mRNA [80].
iii) Alternative Poly(A) Site Selection
With regard to alternative poly (A) site selection (APA), the SR protein XL6 has largest number of CR-APA (coding region alternative poly (A) site selection) events and Rbp1 has least number of events. In most of the cases (either CR-APA or 3’UTR APA), reduction of SR proteins leads to preference of proximal site usage over the distal site usage except a few cases such as B52 and SC35 where many more events resulted in distal site usage. The CR-APA may interact with spliceosome components, but 3’UTR-APA acts independent of splicing events and may implicate SR protein interaction with the 3’ processing complex [1].
In our study, the ESE distributions are gene specific and the smaller the gene the more abundance of ESEs exists. The consensus patterns are the abundance of SF2/ASF in the 1st exon in comparison with the last exon and SRp40 and SRp55 are more shifted toward the 3’ side of the gene. These findings suggest that the functions of SF2/ASF and SRp40/SRp55 may be different from simple splicing enhancement. SF2/ASF has been reported to have suppressive effects on R- loop formation for the DNA stabilization.
4.1.3 ESEs on mRNA Export, Localization, Translation and Non-sense Mediated Degradation (NMD)
A subset of SR proteins shuttle between the cell nucleus and the cytoplasm. These include SF2/ASF, 9G8, SRp20 and others. These SR proteins bind to TAP, which is an mRNA export receptor, by its N-terminal domain and is involved in export of intron spliced mRNA as well as non-intron containing mRNA [7]. The SF2/ASF, in the cytoplasm, is associated with polysomes and stimulates translation. Using a gene construct containing EDA ESE, which is recognized by SF2/ASF and 9G8, it was observed that reporter gene expression is stimulated by SF2. On the other hand, in the gene constructs containing ESE motifs for SRp20 or SC35, there was no enhancement of translation by SRp20 or SC35 [6]. The SRp40 and SRp55 motifs have stimulatory effects on long insulin chain mRNA translation [35, 36].
The shuttling of SR proteins is regulated by phosphorylation of the SR domain in these proteins. The phosphorylated SR proteins enter the nucleus. On the other hand, the dephosphorylated SR proteins exit the nucleus [81]. Some shuttle freely and some carry mRNA with them into the cytoplasm. The SR proteins that function as export adapters include SRp20 and 9G8. These SR-proteins bind to the histone H2a mRNA export element and enhance mRNA export [82].
4.2 ESEs in Non-coding RNA
The cross-linking and immunoprecipitation by SR protein antibodies, followed by high-throughput sequencing (iCLIP-seq), revealed that the SR protein binds not only to intron containing pre- mRNA but also to diverse classes of RNAs including intronless pre-mRNA and non-coding RNAs which are snRNA, tRNA, snoRNA, lncRNA and others. These facts implicate the ESE’s function in extra biological reactions other than in splicing. The global landscape shows the clustering of ESEs in exons and introns of any intron containing pre-mRNAs more than 5’UTR, and the least is found in 3’ UTR. Among non-coding RNAs, the high ESE clustering is present in snoRNA and tRNA as well as other non-coding RNAs. The clustering of ESE in rRNA, snRNA and miRNA is low in comparison with other non-coding RNAs [1].
In addition, we examined the distribution of ESEs in lncRNA as well as DNA breakpoints regions. The distribution of specific ESEs in different ncRNAa was revealed where Alu RNA was high in abundance of ESEs with SF2/ASF domination. The cancer related lncRNA MALAT1 and DNA breakpoints have relatively less ESEs but shows SRp40 domination. The ESEs in satellite DNAs have a varying number of ESEs in the order of α (~12/100 nt) <β (~16.5/100 nt) <γ (~22/ 100 nt) in abundance.
Satellite III has ~ an equal number of ESEs as α-satellite. Whether the ESE has any role in transposon activity is not yet known but the abundance in small genes and Alu elements suggest that they may play a role. The presence of specific ESE motifs in triplet repeats is interesting in view of the alterations in splicing by sequestration of splicing factors and co-factors [73].
Some of the known extra activities of ESEs are as follows:
- SF2 and SC35 have a role in maintenance of DNA stabilities by preventing mutagenic R- loop formation, persistent R-loop, and hypermutation [2].
- SF2 and SC35 also enhance transcription by recruiting p-TEFb and other transcription factors to the transcription complex site. Depletion of SF2/ASF or/and SC35 decrease transcription activity and SC35 enhances transcriptional elongation in a gene specific manner. Deletion of SC35 leads to accumulation of pol II on gene bodies [3]
- Involved in export, localization, translation and nonsense mediated decay (NMD)
- Involved in miRNA biogenesis
SRSF1 (SF2/ASF) and SRSF3 (SRp20) are considered as oncogenes because they are expressed highly in tumor cells such as human U20S and in HeLa cells. Knock-down of these SR-proteins prevents cell proliferation of these cells [83]. These SR proteins work alone at one ESE as well as in combinations with ESEs. They also have inter-relations between SR-proteins. Overexpression of SRp20 increases SF2 expression and overexpression of SF2 increases SRp20 expression. The knock-down or overexpression of SRp20 affect many cell cycle control proteins in alternative splicing and expression.
In mice, the SRp20 gene has 7 exons and its exon 4 is alternatively spliced depending on the nutritional state of the cells. Under fed condition, exon 4 is skipped producing a full length protein and under starved condition, the exon 4 is included and produces a truncated protein without a SR domain at the C-terminus [84].
Satellite DNA is a component of heterochromatin at the centromere and telomere regions. Some of the sequences are well conserved and some have stochastic mutations which differ from species to species. The α-satellite is composed of ~170 nucleotides of repeat sequences or contains oligonucleotide (pentanucleotides) repeat elements. It has a specific protein binding domains such as the CENP A or CENP B binding domains. It is also transcriptionally active, producing siRNA precursors or ribozymes, as part of a 5’ or 3’ UTR mRNA transcript. Their transcriptional activity depends on development, cell types and stress conditions [85]. The inactive genes in heterochromatin have histone markers specific to that locus.
The Histone H3 lysine 9 methylation in C. elegans is generally a repressive modification on transcription. These H3K9me2 or H3K9me3 are enriched in tissue specific silent genes and repetitive elements. The H3K9me2 or H3K9me3 modifications stabilize and protect repeat rich genomes by suppressing transcription induced replicative stress. In met-2 set-25 double mutants, transposons and simple repeats are de-repressed in germline and somatic tissues, leading to increased repeat specific insertions, deletions, copy number variations, R loops and enhanced sensitivity to replicative stress [86].
Table 2a. ESEs in Gene Transcripts (Number of ESEs/100 nt)
|
|
FMR1 |
Ovomucoid |
25VD3H |
APRT (Hams) |
Insulin |
|
Total length |
10.53 (39,224nt) |
14.59 (6,067nt) |
17.14 (4,825nt) |
17.02 (2,251nt) |
19.97 (1,430nt) |
|
Exons |
10.96 (4,456nt) |
15.80 (1,424nt) |
17.03 (2,551nt) |
17.42 (881nt) |
20.64 (465nt) |
|
Introns |
10.47 (34,768nt) |
15.22 (4,643nt) |
17.20 (2,274nt) |
I6.75 (l,370nt) |
19.64 (965nt) |
|
Splice Sites |
9.82 (3,226nt) |
14.90 (1,400) |
18.42 (1,569nt) |
18.07 |
23.72 (392nt) |
The ESE motifs are scanned by ESE finder 3 (Cold Spring Harbor Laboratory) and counting the number of ESEs above threshold score designated in the program. The threshold values are SF2/ASF (1.956); SF2/ASF (IgM-BRCA) (1.867); SC35 (2.383); SRp40 (2.67) and SRp55 (2.676).
The total number of ESEs is divided by the total number of nucleotides and multiplied by 100. The values in the Table represent the number of ESEs per 100 nucleotides in different gene transcripts.
Transcript sequences were obtained from NCBI and Ensemble release.
The human insulin gene (NCBI; J00265), hamster APRT (adenine phosphoribosyltransferase) gene (NCBI; X03603), human 25-hydroxyvitamin D3 1-α-hydroxylase gene (NCBI; AB006987), chicken ovomucoid gene (Ensemble release 43, http://www.ensemble.org), and FMR1 (NCBI; L29074.1) (see Materials and Methods).
The shorter the gene, the more ESE abundance is found. The Exons in FMR1 are 11.36%, in ovomucoid they are 23.47%, in 25 hydrxyvitamin D3 1-α hydroxylase they are 52.87%, in APRT they are 39.14% and in insulin they are 32.52%.
Table 2b. Total ESE counts in Gene Transcripts
|
|
SF2/ASF |
SC35 |
SRp40 |
SRp55 |
|
FMR1 (Human) 39,224 nt |
830 |
1,108 |
1,290 |
902 |
|
Ovomucoid (Chicken) 6,067 nt |
227 |
230 |
237 |
190 |
|
25VD3αH (Human) 4,825 nt |
270 |
220 |
194 |
142 |
|
APRT (Hamster) 2,251 nt |
109 |
113 |
107 |
54 |
|
Insulin (Human) 1,430 nt |
115 |
74 |
65 |
45 |
|
Total 53,797 nt |
1,550 |
1,745 |
1,893 |
1,332 |
The numbers of ESEs are counted and summed up for the total number in all gene transcripts (53,797 nucleotides). Numbers in the SF2/ASF column represent an average of SF2/ASF (1.956) and SF2/ASF (IgM-BRCA) (1.867). Numbers in parenthesis are threshold values for each motif. Each individual gene has its characteristic ESE content but overall, the SRp40 is the most abundant and SRp55 is the least abundant in this group of coding genes.
Table 2c. Individual ESEs in Gene transcripts (Number of ESEs/100 nt)
|
Gene |
SF2/ASF |
SC35 |
SRp40 |
SR55 |
Σ |
|
FMR1 (39,224 nt) |
2.12 |
2.82 |
3.29 |
2.30 |
10.53 |
|
Exon (4,456 nt) |
3.18 |
2.60 |
3.21 |
1.97 |
10.96 |
|
Intron (34,768 nt) |
1.98 |
2.85 |
3.30 |
2.34 |
10.47 |
|
SS (3,226 nt) |
2.26 |
2.11 |
3.50 |
1.95 |
9.82 |
|
|
|||||
|
Ovomucoid (6,067 nt) |
3.76 |
3.79 |
3.91 |
3.13 |
14.59 |
|
Exon (1,424 nt) |
4.57 |
4.28 |
3.72 |
3.23 |
15.80 |
|
Intron (4,643 nt) |
3.52 |
4.64 |
3.96 |
3.10 |
15.22 |
|
SS (1,400 nt) |
4.47 |
3.36 |
3.57 |
3.50 |
14.90 |
|
|
|||||
|
25HVD3H (4,825 nt) |
5.62 |
4.56 |
4.02 |
2.94 |
17.14 |
|
Exon (2,551 nt) |
5.47 |
4.78 |
3.80 |
2.98 |
17.03 |
|
Intron (2,274 nt) |
5.72 |
4.31 |
4.27 |
2.90 |
17.20 |
|
SS (1,569 nt) |
6.57 |
5.35 |
4.40 |
2.10 |
18.42 |
|
|
|||||
|
APRT (2,251 nt) |
4.84 |
5.02 |
4.75 |
2.40 |
17.02 |
|
Exon (881 nt) |
5.39 |
5.22 |
3.63 |
3.18 |
17.42 |
|
Intron (1,370 nt) |
4.49 |
4.89 |
5.47 |
1.90 |
16.75 |
|
SS (800 nt) |
5.32 |
5.00 |
5.50 |
2.25 |
18.07 |
|
|
|
|
|||
|
Insulin (1,430 nt) |
7.10 |
5.17 |
4.55 |
3.15 |
19.97 |
|
Exon (465 nt) |
6.45 |
4.73 |
5.16 |
4.30 |
20.64 |
|
Intron (965 nt) |
7.41 |
5.39 |
4.25 |
2.59 |
19.64 |
|
SS (392 nt) |
7.65 |
7.14 |
5.61 |
3.32 |
23.72 |
|
|
|||||
|
Av. 5 genes; Total |
4.69 |
4.31 |
4.10 |
2.78 |
15.85 |
|
Exon |
5.01 |
4.41 |
3.78 |
3.13 |
16.37 |
|
Intron |
4.62 |
4.42 |
4.25 |
2.57 |
15.86 |
|
SS |
5.25 |
4.59 |
4.52 |
2.62 |
16.99 |
The numbers of ESE in 100 nucleotides are calculated in total sequences, exons only, introns only and splice sites. The splice sites include 200 nucleotides at each splice site which include 100 nucleotides on the 5’ splice site and 100 nucleotides on the 3’ splice site. Numbers in the SF2/ASF column represent an average of SF2/ASF (1.956) and SF2/ASF (IgM-BRCA) (1.867).
ESE counts are made above the default threshold value stated in Table 2a. It is evident that splice sites have more ESE cluster than other sites in the order of SS>Exon>Intron. However, there are gene specific differences. It is interesting to note that SC35 and SRp40 are more abundant in introns than in exons.
Table 3a. ESE Ratio 1st/last
|
Gene |
SF2/ASF |
SC35 |
SRp40 |
SRp55 |
Σ |
|
Insulin |
1.36 |
1.16 |
1.74 |
0.47 |
1.16 |
|
APRT |
2.45 |
1.46 |
0.58 |
0.84 |
1.36 |
|
25VD3H |
2.68 |
1.41 |
1.55 |
1.04 |
1.66 |
|
Ovomucoid |
1.78 |
0.69 |
1.19 |
1.66 |
1.26 |
|
FMR1 |
9.17 |
1.94 |
1.21 |
1.19 |
2.86 |
|
Σ |
2.62 |
1.27 |
1,27 |
0.91 |
1.52 |
Table 3b. ESE Ratio last/1st
|
Gene |
SF2/ASF |
SC35 |
SRp40 |
SRp55 |
Σ |
|
Insulin |
0.74 |
0.86 |
0.58 |
2.11 |
0.8 |
|
APRT |
0.41 |
0.68 |
1.71 |
1.20 |
0.74 |
|
25VD3H |
0.37 |
0.71 |
0.65 |
0.96 |
0.60 |
|
Ovomucoid |
0.5 |
1.45 |
0.84 |
0.60 |
0.79 |
|
FMR1 |
0.11 |
0.51 |
0.83 |
0.84 |
0.35 |
|
Σ |
0.38 |
0.79 |
0.79 |
1.09 |
0.66 |
The numbers of ESEs are counted and calculated as numbers of ESEs per 100 nucleotides. The ratio of the first exon to the last exon is the product of division of numbers of ESEs in the first exon by the numbers of ESEs in the last exon.
The ratio of last to first is also calculated accordingly. It is evident that the ratio of 1st/last is highest in SF2/ASF and is above 1 in all cases, indicating clustering of SF2/ASF in first exons and an abundance of sum of ESEs (Σ column) in first exons are higher than in the last exons. The overall abundance of ESEs in last exons are fewer but the SRp55 is relatively more frequent than in the first exon.
Table 4. SR protein Binding Sites per 100 nucleotides in RRE and CTE
|
RNA |
SF2/ASF |
SC35 |
SRp40 |
SRp55 |
Σ |
|
RRE (240 nt); Default |
5.42 |
3.75 |
3.75 |
4.58 |
17.5 |
|
Above 0 |
16.04 |
23.3 |
25.0 |
17.92 |
82.3 |
|
CTE (235 nt); Default |
6.17 |
3.83 |
5.53 |
2.55 |
18.09 |
|
Above 0 |
20.64 |
21.28 |
24.68 |
16.17 |
82.77 |
The SR protein binding sites are screened by ESEfinder 3 (CSHL). The default threshold values are: SF2/ASF (1.956); SF2/ASF (IgM-BRCA) (1.867); SC35 (2.383); SRp40 (2.67) and SRp55 (2.676). The numbers of SF2/ASF in the table are the averages of SF2/ASF and SF2/ASF (IgM- BRCA). Those above 0 were counted for each SR protein binding motif. At the default threshold, SF2 is the most abundant motif and above 0 counts the SRp40 is the most abundant motif. Accordingly, the translation promoting activity of introns containing genomic RNA are motif specific rather than being due to their abundance.
The RRE sequence is from Daugherty et al., [87]. The CTE sequence is from Rizvi et al., [88]
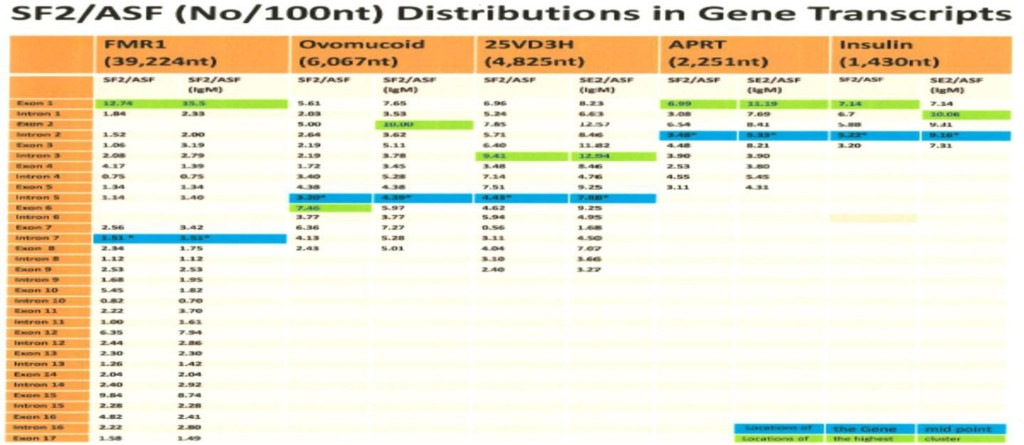
Table 5. ESE Distribution (ESE/100 nucleotides) in FMR1 Gene Transcript
|
FMR1 Gene |
SF2/ASF |
SC35 |
SRp40 |
SRp55 |
Σ |
|
Total (39,224 nt) |
2.12 |
2.82 |
3.29 |
2.30 |
10.53 |
|
Exons (4,456 nt) |
3.18 |
2.60 |
3.21 |
1.97 |
10.96 |
|
Introns (34,768 nt) |
1.98 |
2.85 |
3.30 |
2.34 |
10.47 |
|
SS (3,226 nt) |
2.26 |
2.11 |
3.50 |
1.95 |
9.82 |
|
Alu (2,244 nt) |
4.93 |
5.21 |
4.32 |
1.34 |
15.80 |
|
LINE (211 nt) |
4.74 |
2.37 |
2.84 |
4.27 |
14.22 |
|
Σ |
19.21 |
17.96 |
16.96 |
14.17 |
|
The FMR1 gene is typical of hnRNA with only 11.36% exons, but the ESE distribution is different from other shorter pre-mRNAs with high a proportion of exons. However, the Alu, LINE and other short repeating sequences are clustered with ESEs.
Table 6. ESEs /100 Nucleotides at Splice Sites
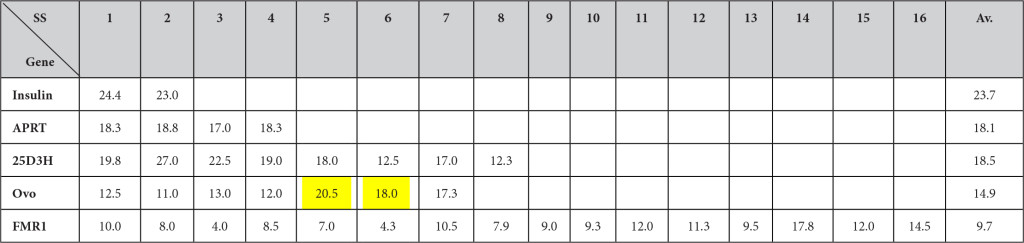
The numbers of ESEs at splice sites were counted 50 nucleotides upstream from GU and 50 nucleotides downstream from the G at GU for 5’ splice sites and 50 nucleotides upstream from G at AG and 50 nucleotides downstream from AG from the 3’ splice sites. If exons or introns were shorter than 50 nucleotides, they includde entire exons or introns without extending further. The numbers of ESEs then were calculated per 100 nucleotides. It is evident that the high clustering at splice sites is seen in shorter genes such as the insulin gene, but the trends are fading when genes become longer. The yellow highlighted sites in ovomucoid genes indicate where the splicing is taking place much faster (or earlier) than other sites.
Table 7. ESE Distribution at 5’ and 3’ Splice Sites, ESEs/100 Nucleotides at Splice Sites
|
Gene |
SF2/IgM (Average) |
SC35 |
SRp40 |
SRp55 |
||||
|
Insulin (Human) |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
|
7.81 > 7.50 |
7.29 > 7.00 |
7.81 > 3.50 |
2.08 < 4.50 |
|||||
|
APRT (Hamster) |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
|
6.00 > 4.63 |
4.50 < 5.50 |
4.50 < 6.50 |
3.00 > 1.50 |
|||||
|
25VD3H (Human) |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
|
6.63 > 6.50 |
5.88 > 4.81 |
4.13 < 4.68 |
1.88 < 2.34 |
|||||
|
Ovomucoid (Chicken) |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
|
4.00 < 4.99 |
3.71 > 3.00 |
3.14 < 4.00 |
4.57 > 2.43 |
|||||
|
FMR1 (Human) |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
|
2.47 > 2.06 |
3.5 > 1.85 |
3.13 < 3.87 |
1.88 < 2.03 |
|||||
|
Average (5 genes) |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
5’ SS |
3’ SS |
|
5.38 > 5.14 |
4.98 > 4.43 |
4.54 > 4.51 |
2.68 > 2.56 |
|||||
The numbers of SESs were determined at the 5’ and 3’ splice sites. The 100 nucleotides at the 5’ splice sites include 50 nucleotides of exon and 50 nucleotides from G at GU extend into introns. The 100 nucleotides at the 3’ splice sites include intron 50 nucleotides up to AG and 50 nucleotides in exon. The clustering is close to even between 5’ splice sites and 3’ splice sites, but in most cases (average of all 5 genes combined), a little more is evident at the 5’ splice sites.
Table 8. ESEs in Alu Elements (Number of ESEs in 100 nucleotides)
|
RNA |
SF2/ASF |
SC35 |
SRp40 |
SRp55 |
Total |
5’SS |
3’SS |
Brnc. S |
|
FMR1 Alu(+)(1,371 nt) Alu(-) (873 nt) Average |
5.30 4.31 4.93 |
4.62 6.19 5,21 |
3.89 5.04 4.32 |
1.17 1.61 1.34 |
14.98 17.11 15.80 |
3.61 2.20 3.08 |
2.97 4.58 3.58 |
9.51 13.8 11.1 |
|
Consensus Alu Major (120 nt) Precise (118 nt) PV(HS) (118 nt) ∑ (356 nt) |
9.2 7.6 6.8 7.6 |
4.2 5.1 5.1 4.8 |
3.3 4.2 4.2 3.9 |
0.8 0.8 0.8 0.8 |
17.5 17.8 16.9 17.1 |
3.3 3.4 3.4 3.4 |
1.7 0.8 0.8 1.1 |
5.8 5.1 5.1 5.3 |
|
hYRNA (389 nt) |
3.6 |
4.3 |
5.6 |
1.0 |
14.7 |
1.5 |
1.0 |
9.0 |
|
Fish SB (696 nt) |
4.3 |
3.6 |
3.7 |
2.6 |
14.2 |
1.7 |
1.1 |
8.9 |
|
Rat 4.5S RNA I (96 nt) |
3.65 |
3.13 |
6.25 |
1.04 |
14.07 |
0 |
2.08 |
10.4 |
|
Σ Average |
5.60 |
4.53 |
4.52 |
1.23 |
15.91 |
2.39 |
1.88 |
8.45 |
The numbers of individual ESEs are counted in total numbers of nucleotides in a class of Alu RNAs and calculated numbers of ESEs per 100 nucleotides. The consensus is the total number of ESEs which is in the range of 15–18 per 100 nucleotides The SF2/ASF appears to dominate most of the Alu elements (Σ Average) while SRp40 is dominating in YRNA and rat 4.5S RNAI. The Alu (-) has SC35 domination over SF2/ASF. The Alu (+) has more 5’ splice sites than 3’ splice sites.
- The Alu in FMR1 is from NCBI GenBank; L29074.1
- The Consensus Alu sequences are from Maraia et al., [29]
The Y RNA sequences are from Christov et al., [30]. - The sequences of SB (sleeping beauty) are from Hackett et al., [31], Ivics et al., [89] and van Pouderoyen et al., [32]
- The 4.5S RNA sequence is from Ro-Choi et al., [90]
Table 9. ESEs/100 nt in DNA Breakpoints, MALAT1 and NEAT1
|
Nucleic Acids |
SF2/ASF |
SC35 |
SRp40 |
SRp55 |
∑ |
|
DNA Breakpoints (total 5,020 nt) (H) Hybrid (1,610 nt) (+) Strand (1,716 nt) |
2.6 2.6 2.9 2.5 |
2.9 2.8 2.7 3.2 |
3.5 3.4 3.7 3.5 |
2.0 2.4 1.9 1.7 |
11.0 11.0 11.0 11.0 |
|
NEAT1 (3,756 nt) |
3.7 |
2.9 |
3.1 |
2.1 |
11.8 |
|
MALAT1 (8,758 nt) |
2.7 |
2.9 |
3.6 |
1.9 |
11.1 |
The numbers of ESEs were counted above the default thresholds which are:
SF2/ASF (1.956); SF2/ASF (IgM-BRCA) (1.867); SC35 (2.383); SRp40 (2.67) and SRp55 (2.676) and calculated for the numbers of ESEs per 100 nucleotides. In this group of sequences, the SRp40 is dominating over other ESE motifs.
- The DNA break points are from Liu et al., [26] and Chen et al., [91].
- The NEAT1 sequence is from NCBI; NR_028272.1
- MALAT1 is from NCBI; NR_002819.3.
Table 10. ESEs in Satellite DNAs (Numbers of ESE per 100 nucleotides)
|
DNA |
SF2/ASF |
SC35 |
SRp40 |
SRp55 |
∑ |
|
α-Satellite Consensus 1 (171 nt) 2 (170 nt) 3 (171 nt) 4 (171 nt) 5 (169 nt) Average |
2.92 2.94 5.26 5.26 3.55 3.99 |
0.58 1.18 1.75 1.17 1.18 1.17 |
2.92 1.76 5.85 2.34 3.37 3.05 |
3.51 3.51 2.92 4.09 3.55 3.52 |
9.93 9.39 15.78 12.86 10.65 11.73 |
|
Chromosome 17 α-Satellite |
4.46 |
0.97 |
3.48 |
2.65 |
11.56 |
|
Alphoid (334 nt) |
4.19 |
1.50 |
4.19 |
3.29 |
13.17 |
|
β-Satellite Acrocentric chromosome (955 nt) |
5.13 |
3.66 |
4.61 |
3.46 |
16.86 |
|
Chromosome 9p12 β-Satellite (69 nt) |
7.25 |
2.90 |
2.90 |
2.90 |
15.95 |
|
γ-Satellite (1,962 nt) |
8.46 |
5.86 |
5.15 |
2.09 |
21.56 |
|
Satellite III Chromosome 14 (1,404 nt) |
2.42 |
1.42 |
2.35 |
1.14 |
7.33 |
|
Satellite III Chromosome 9 |
5.06 |
3.16 |
3.86 |
0.63 |
12.71 |
The counting and calculations are same as in other tables.
There are wide ranges of ESE motifs in different classes of satellite DNA. However, the SF2/ASF appears to dominate in its abundance.
- The human α-satellite consensus 1 (chromosome 20) is from NCBI, GenBank L06776.1
- The human α-satellite consensus sequences 1 and 2 are from Waye and Willard, (1987). [92]
- The human α-satellite consensus sequence 3, 4 and 5 are from Vissel and Choo, (1987). [93]
- The human α-satellite consensus 4 (chromosome 4) is from NCBI, GenBank S67971.1
- The α-satellite repeat from human chromosome 17 (718 bp) is from NCBI GenBank; L08550.1 The human alphoid (334 bp) is from NCBI, GenBank S49988.1
- The β-satellite sequence (955 nt) is from NCBI, GenBank M81228.1(Acrocentric chromosome)
- The β-satellite sequence (69 nt) in chromosome 9 is from NCBI, GenBank M25748.1
- The human γ-satellite sequence (1,962 nt) is from NCBI, GenBank; X68546.1 (Chromosome 8)
- The satellite III sequence (1,404 nt) is from NCBI GenBank; S90110.1 (Chromosome 14)
- The satellite III sequence (158 nt) in chromosome 9q11-q12 is from Jolly et al., [94]
Table 11. ESE Clusters in Repeat Sequences
|
Repeats |
SF2/ASF |
SF2/ASF(lgM) |
SC35 |
SRp40 |
SRp55 |
|
CGG Repeats |
34/100 |
34/100 |
0 |
0 |
0 |
|
CAG Repeats |
0 |
0 |
0 |
0 |
34/100 |
|
CUG Repeats |
0 |
0 |
34/100 |
0 |
34/100 |
|
CCUG Repeats |
1.38/1.956 |
26/100 |
O |
0 |
0 |
|
AUUCU Repeats |
0 |
O |
O |
0 |
0 |
The repeat sequences are from Mirkin, S.M. [33]
Repeat sequences are subjected to the ESEfinder3 and the selected motifs are above default threshold values. Numbers of motifs are calculated in 100 nucleotide bins. The SF2/ASF in CCUG, the positive motifs are at the +1.38, while threshold value is +1.956. At the +1.38, there are 26 motifs of SF2/ASF.
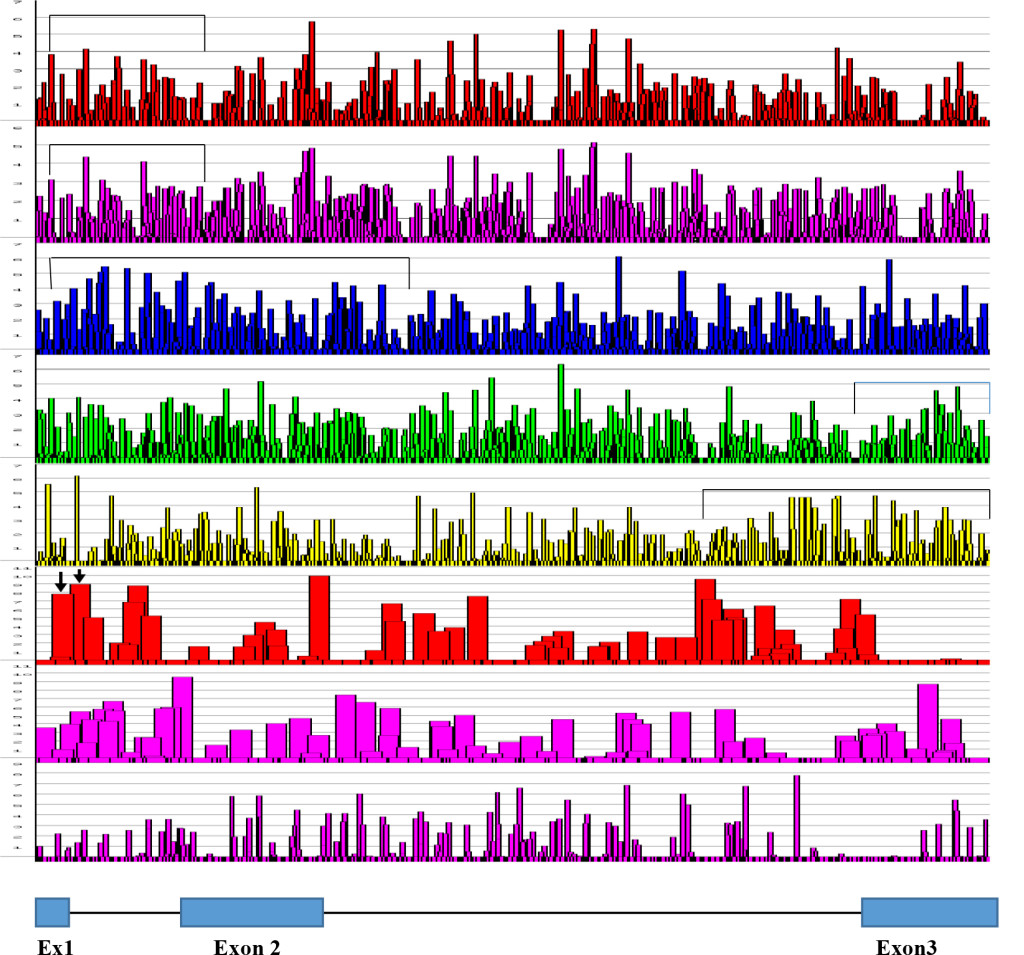
Figure 1: ESE Distribution in Human Insulin Gene Transcript [From the Top; SF2/ASF, SF2/ASF(IgM), SC35, SRp40, SRp55, 5’SS, 3’SS, Br. S]
The human Insulin gene sequence was retrieved from NCBI J00265 and subjected to ESE screening by ESEfinder 3, Cold Spring Harbor Laboratory.
Graphical presentation is from the ESE finder 3, CSHL and clustered regions are bracketed by ![]()
SF2/ASF clustering is in the 5’ side of the gene, SC35 is more in the gene body with some on the 5’ side. SRp40 and SRp55 are clustered more on 3’ side of the molecule. Arrows in the 5’SS scan represent splice site at nucleotide position at 42 and alternative splice site at nucleotide position at 68.
The structural organization of insulin gene is as follows:
- Exon 1 is from nucleotide 1 to 42 (42 nucleotides)
- Intron 1 is from nucleotide 43 to 221 (179 nucleotides)
- Exon 2 is from nucleotide 222 to 425 (204 nucleotides)
- Intron 2 is from nucleotide 426 to 1,211 (786 nucleotides)
- Exon 3 is from nucleotide 1,212 to 1,430 (219 nucleotides)
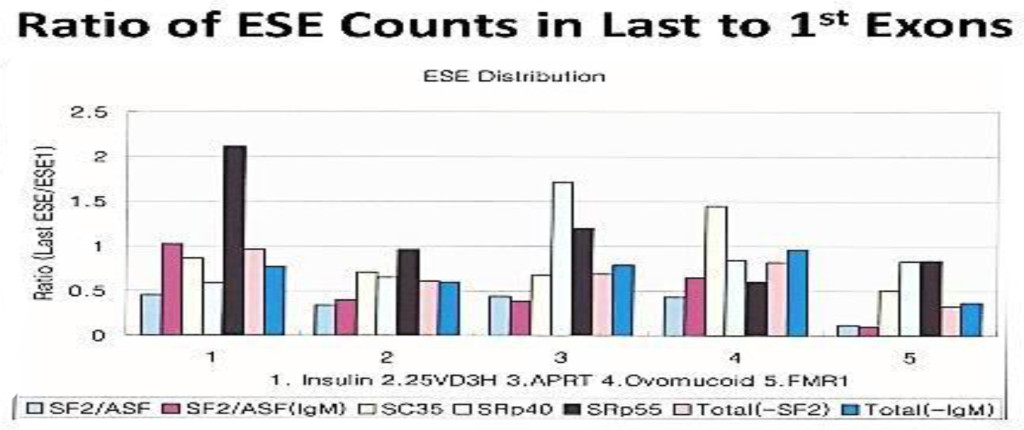
Figure 2. Ratio of ESE counts in last to 1st Exons
The distribution of ESEs are gene specific. However, there are some consensus patterns;
the SF2/ASF is clustered more at in the 1st exon than the last exon; SRp40 and SRp 55 are more clustered at the last exon than the 1st.
Figures 3a
Figures 3b
Figures 3c
Figures 3d
Figure 3. Distribution of ESE in Exons and Introns. The numbers of ESEs are presented in each 100 nucleotide bin. The blue marks represent the middle exon/intron and green marks represent the highest ESE containing exon/intron. The distribution appears to be even throughout the molecule with some focal clustering. The highest cluster of SF2/ASF (9 out of 10) and SC35 (4 out of 5) are in the 5’ side of the molecule and the highest cluster of SRp40 (3 out of 5) is in 3’ side of the molecule. However, the first exons contain the SF2 abundance; SC35 is more in the gene body; SRp40 and SRp55 are clustered toward to the 3’ side of the molecule.
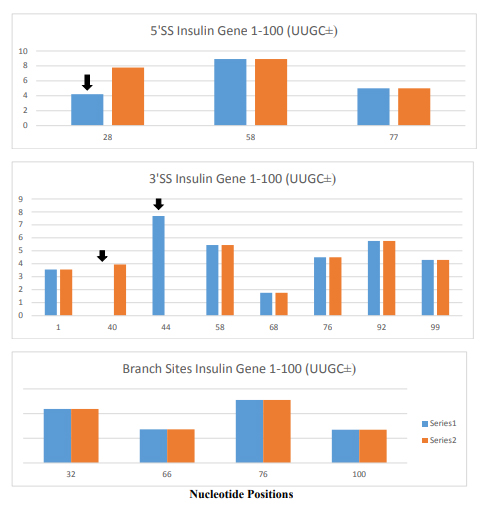
Figure 4. Changes in 5’ Splice Sites, 3’ Splice Sites and Branch Sites in Insulin Gene Variant (IVS-69). The insulin gene variant (IVS-69) has TTGC insertion at nucleotide position 47 to 50. The blue columns are the normal insulin gene and orange columns are for the insulin gene variant. The X- axis is the position of nucleotide in insulin gene and Y-axis is the score of strength at the splice sites.
The changes in 5’ splice site such as attenuation of 5’ splice site at the position 28 (marked by black arrow) alters 5’ splice site usage at position 58 producing 30 nucleotides longer 5’ UTR containing insulin mRNA. There are changes in 3’ splice sites (marked by black arrows) as well, but its significance is not known. Its close proximity to 5’ splice site suggests that it may interfere the formation of spliceosome at the canonical 5’ splice site. There was no change in branch site by the insertion of TTGC insertion.
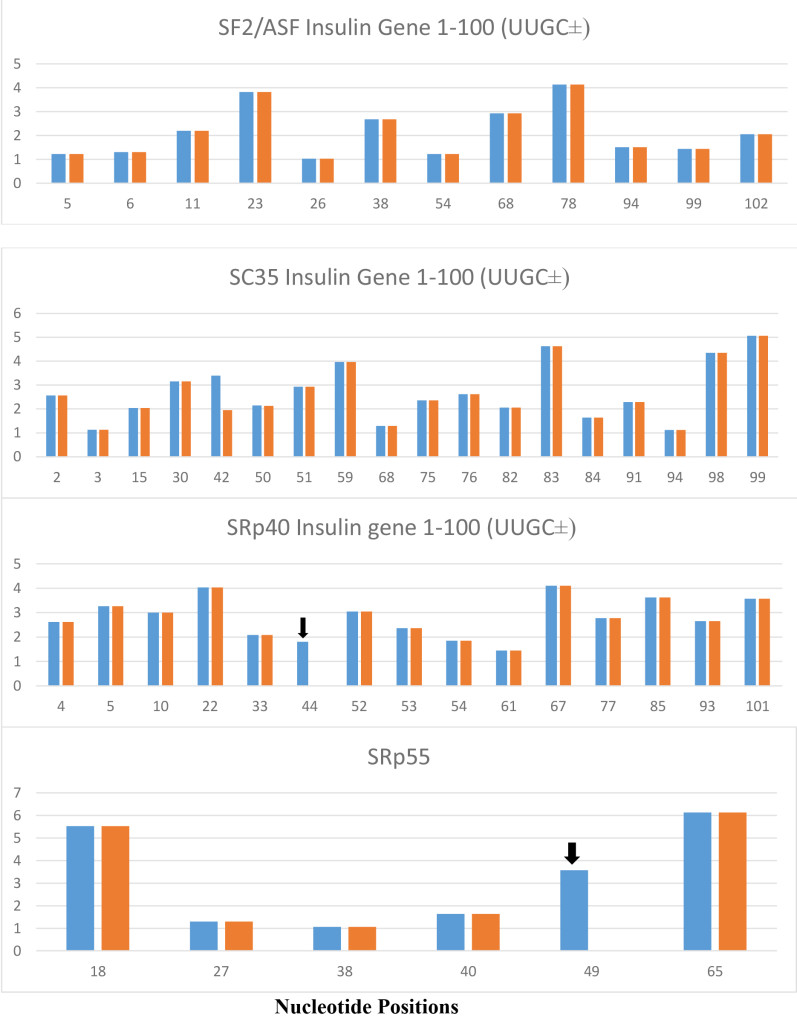
Figure 5. The variant insulin gene IVS-69, which contains UUGC insertion at a position 6 nt downstream from 5’ splice site of intron 1 (position 47–50 from TSS), is present exclusively in Africans and produces variant insulin mRNA with extended 5’ UTR. The UUGC insertion produces additional SRp40 at position 44 and additional SRp55 at the position 49 which are indicated by black arrows. The SRp40 and SRp55 co-expression with reporter insulin pre-mRNA construct increased the proportion of transcript retaining intron 1 and increased proinsulin level in the cell [35]. There are no changes in SF2/ASF and SC35.
The blue columns are from normal insulin gene and orange columns are from insulin gene variant (IVS-69). Numbers in X-axis represent the positions of nucleotide in insulin gene and the numbers in Y-axis represent the score of strength of ESE motifs.
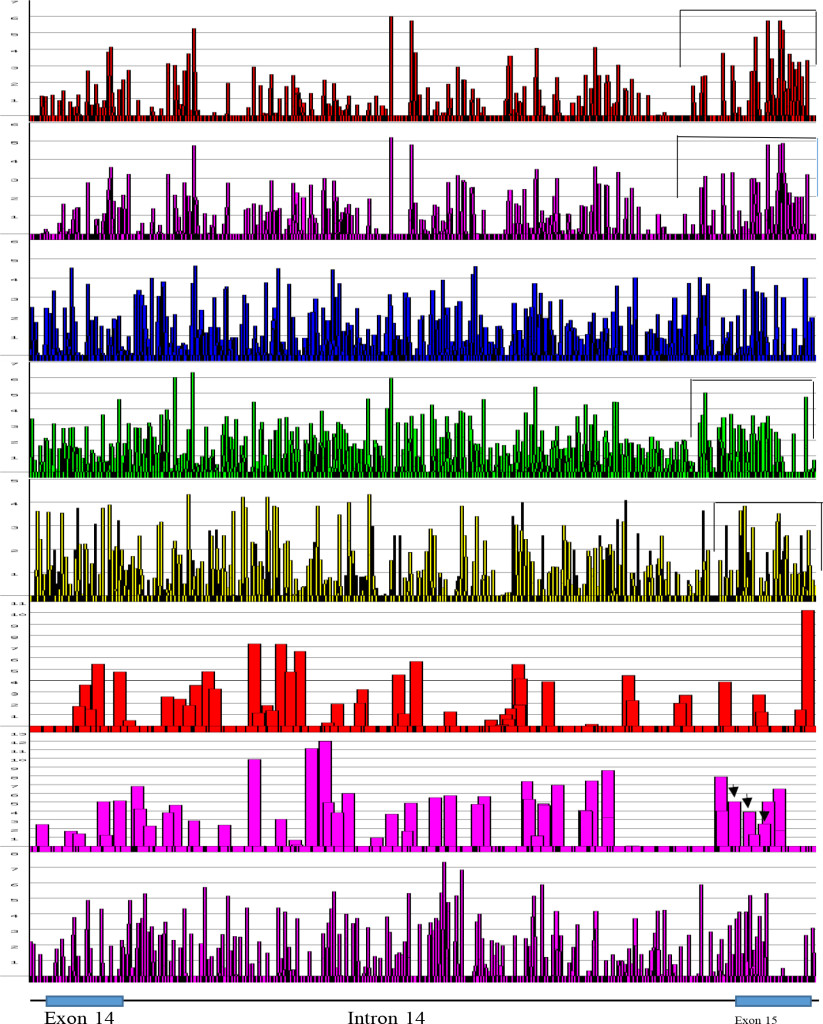
Figure 6. ESEs in FMR1 Exon 14 to Exon 15 at Alternative Splice Sites [From the Top; SF2/ASF, SF2/ASF(IgM), SC35, SRp40, SRp55, 5’SS, 3’SS, Br. S.]
The regions, where the alternative splicings occur were screened for their distribution of ESEs. The 5’ end of exon 15 (FMR1 gene) has two more alternative splice sites in addition to canonical splice site. It is evident, that at the 5’ side of exon 15, there are clustering of SF2/ASF, SRp40 and SRp55. Alternative splice sites are marked by black arrows
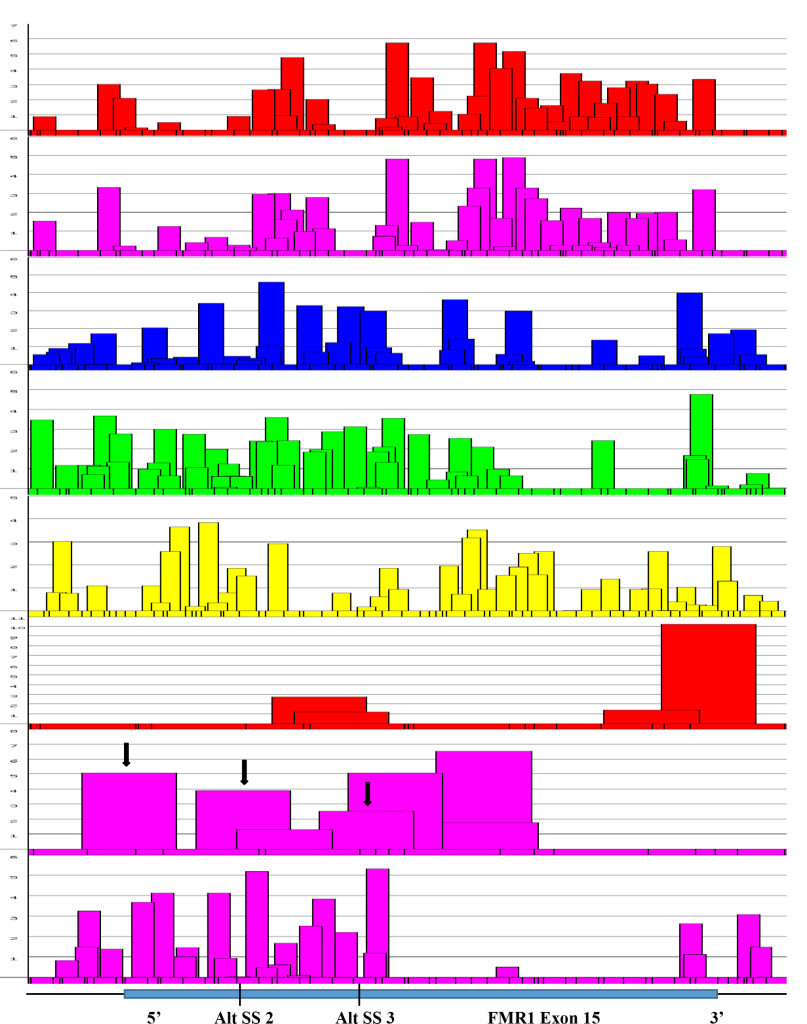
Figure 7. Focal Magnification of Alternative Splice Sites in Exon 15 (FMR1 Gene).
Figure 7. An expanded view of alternative splice sites at exon 15 of FMR1 gene. It was found that the presence of 3’ splice sites at the positions were also where alternative splicings were found (Black arrows). The score of 3’ splicing sites correlated well with the amount of spliced product formed. The canonical site splicing product is the most abundant; next is Alt. SS 2; the least is at the Alt. SS 3. The presence of high scored 3’ splice site next to Alt. SS 3 is not operational. The reason may be due to the presence of splicing silencers at the region (see Figure 8).
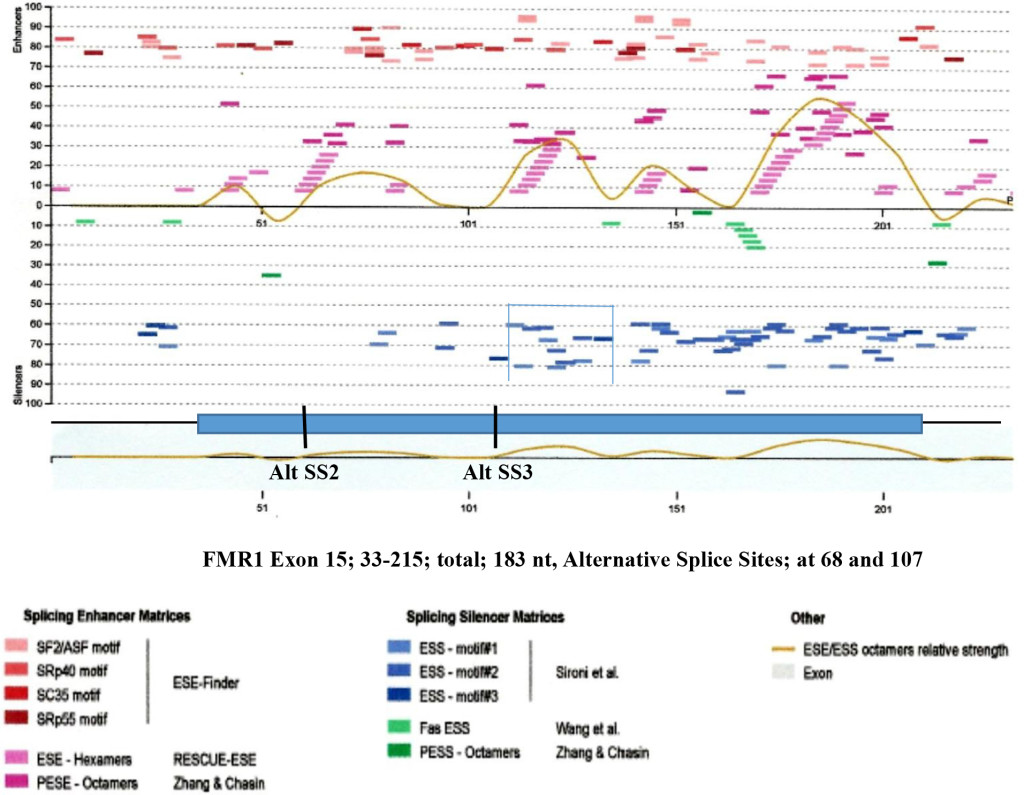
Figure 8. Enhancers and Silencers at Exon 15 (FMR1 Gene).
Figure 8. The splicing enhancers and silencers are scanned by HSF3 [95]. It is interesting to observe that where Alt. SS 2 and Alt. SS 3 are located, there are the least splicing silencer motifs. However in adjacent region, abundant silencer motifs are present (bracketed). The presence of silencer elements may effect enhancers by making them non-operational. Where the clustered silencers are present is marked by blue and green colors
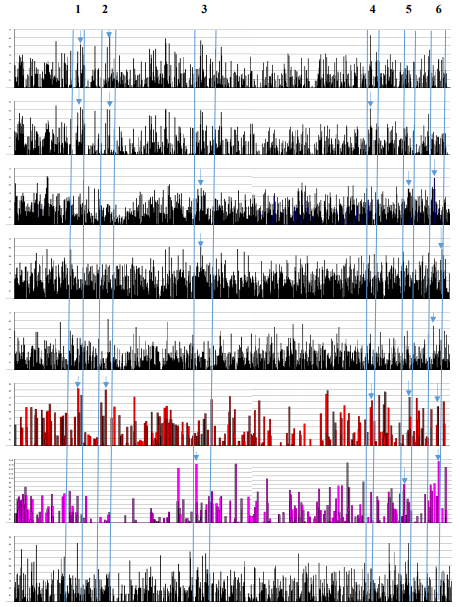
Figure 9. ESE Distributions in MALAT1.
Figure 9. The transcript of the MALAT1 gene at chromosome 11q13.1 (NCBI; NR_002891) has 8,779 nucleotides and contains numerous ESEs and poly A sites. The ESE distributions are from the top to bottom, SF2/ASF, SF2/ASF (IgM-BRCA), SC35, SRp40 and SR55. As in other coding pre- mRNA, SF2/ASF is clustered around the 5’ region, SC35 around the central region and SR-40 and SRp55 are clustered toward the 3’ side. In this graph, ESEfinder 3(CSHL) was used to produce the figure. The 8,779 nucleotides is divided into two portions and a composite graph extending from nucleotide 1–8,779 was constructed.
Although, mature MALAT1 RNA contains a triple helix 3’ end, there are many 5’ and 3’ splice sites as well as poly (A) sites present in the sequence. According to NCBI AceView, the MALAT1 gene can produce at least an additional 10 RNA splices which may or may not have translation products. It is interesting to observe that some of ESE clusters are in correspondence with 5’ or 3’ splice sites present in the sequence.
Examples are
- In column 1, 2 and 4, SF2/ASF ( ↓ ) are in correlation with the high score 5’ splice sites ( ↓ ),
- In column 3, SC35 and SRp40 clusters are corresponding to high score 3’ splice sites and
- In columns 5 and 6, the SC35, SRp40 and SRp55 are at the region where 5’ and 3’ splice sites are.
These ESEs, splice sites and branch sites may not be operational in normal condition, but during degradation, stress or pathologic condition, they may become operational producing aberrant spliced products.
Acknowledgement: Authors are grateful to Dr. Lynn Yeoman for the English corrections.
Refernces
- Bradley T, Cook ME, Blanchette M (2015) SR proteins control a complex network of RNA-processing events. RNA 21: 75–92. [crossref]
- Li X , Manley JL (2005) Inactivation of the SR Protein Splicing Factor ASF/SF2 Results in Genome Instability. Cell 122: 365–378.
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu X-D (2008) The Splicing Factor SC35 has an Active Role in Transcription Elongation. Nat Struct Molec Biol 15: 819–826.
- Lemaire R, Prasad J, Kashima T, Gustafson J, Manley, JL, Lafyatis R (2002) Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev 16: 594–607.
- Cáceres JF, Screaton GR and Krainer AR (1998) A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev 12: 55–66.
- Sanford JR, Gray NK, Beckmann K, C?ceres JF (2004) A novel role for shuttling SR proteins in mRNA translation. Genes Dev 18: 755–768.
- Huang Y, Gattoni R, Stévenin J, Steitz JA (2003) SR Splicing Factors Serve as Adaptor Proteins for TAP-Dependent mRNA Export Mol Cell 11: 837–843.
- Yeo G, Hoon S, Venkatesh B, Burge CB (2004) Variation in sequence and organization of splicing regulatory elements in vertebrate genes Proc Natl Acad Sci 101: 15700–15705.
- Tran Q, Roesser JR (2003) SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing. Biochemistry 42: 951–957. [crossref]
- Cooper DR, Carter G, Li P, Patel R, Watson JE, Patel NA (2014) Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPAR?2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes 5: 1050–1063.
- Fu XD (2004) Towards a splicing code. Cell 119: 736–738. [crossref]
- Dye MJ, Proudfoot NJ (1999) Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell 3: 371–378. [crossref]
- Cramer P, Cáceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR (1999) Coupling of Transcription with Alternative Splicing: RNA Pol II Promoters Modulate SF2/ASF and 9G8 Effects on an Exon Splicing Enhancer. Mol Cell 4: 251–258.
- Goren A, Ram O, Amit M, Karen H, Lev-Maor G, Vig I, Pupko T, Ast G (2006) Comparative Analysis Identifies Exonic Splicing Regulatory Sequences-The Complex Definition of Enhancers and Silencers. Mol Cell 22: 769–781.
- Sapra A, Änkö M-L, Grishina I, Lorenz M, Pabis M, et al. (2009) SR Protein Family Member Display Diverse Activities in the Formation of Nascent and Mature mRNPs In Vivo. Mol Cell 34: 179–190.
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, et al (2004) Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845. [crossref]
- Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126: 37–47. [crossref]
- Cooper TA, Wan L, Dreyfuss G (2009) RNA and Disease. Cell 136: 777–798.
- López-Bigas N, Audit B, Ouzounis C, Parra G, Guigó R (2005) Are splicing mutations the most frequent cause of hereditary disease? FEBS Letter 579: 1900–1903.
- Liu, H-X, Zhang M, rainer AR (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev 12: 1998–2012.
- Liu, H-X, Chew SL, Cartegni L, Zhang MQ, Krainer AR (2000) Exonic Splicing Enhancer Motif Recognized by Human SC35 under Splicing Conditions. Mol Cell Biol 20: 1063–1071.
- Cartegni, , Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucl Acids Res 31: 3568–3571.
- Kashima T and Manley JL (2003) A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nature Genet 34: 460–463.
- Muro AF, Caputi M, Pariyarath R, Pagani F, Buratti E, et al. (1999) Regulation of Fibronectin EDA Exon Alternative Splicing: Possible Role of RNA Secondary Structure for Enhancer Display. Mol Cell Biol 19: 2657–2671.
- Sen, S, Talukdar, I and Webster, NJG (2009) SRp20 and CUG-BP1 Modulate Insulin Receptor Exon 11 Alternative Splicing. Mol Cell Biol 29: 871–880.
- Liu P, Erez A, Nagamani SC, Dhar SU, Kołodziejska KE, et al (2011) Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903. [crossref]
- Waye JS. Willard HF (1987) Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from human chromosomes. Nucl Acids Res 15: 7549–7569.
- Vissel B, Choo KH (1987) Human alpha satellite DNA-consensus sequence and conserved regions. Nucl Acids Res 15: 6751–6752.
- Maraia RJ, Driscoll CT, Bilyeu T, Hsu K, Darlington GJ (1993) Multiple Dispersed Loci Produce Small Cytoplasmic Alu RNA. Mol Cell Biol 13: 4233–4241.
- Christov CP, Gardiner TJ, Szüts D, Krude, T (2006) Functional Requirement of Noncoding Y RNAs for Human Chromosomal DNA Replication. Mol Cell Biol 26: 6993–7004.
- Hackett PB, Ekker SC, Largaespada DA, Mclvor RS (2004) Sleeping Beauty Transposone-Mediated Gene Therapy for Prolonged Expression In: “Non-viral Vectors for Gene Therapy” 2nd Edition (Eds; Huang, L, Wagner, E and Hung, M-C)
- van Pouderoyen G, Ketting RE, Perrakis A, Plasterk RHA, Sixma TK (1997) Crystal structure of the specific DNA binding domain of Tc3 transposase of C elegans in Complex with transposon DNA. EMBO J 16: 6044–6054.
- Mirkin SM (2007) Expandable DNA repeats and human disease. Nature 447: 932–940. [crossref]
- Shalev A, Blair PJ, Hoffmann SC, Hirshberg B, Peculis BA, et al (2002) A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology 143: 2541–2547. [crossref]
- Královicová J, Gaunt TR, Rodriguez S Wood PJ, Day INM, Vorechovský I (2006) Variants in Human Insulin Gene That Affect Pre-mRNA Splicing. Diabetes 55: 260–264.
- Minn AH, Lan H, Rabaglia ME, Harlan DM, Peculis BA, et al (2005) Increased insulin translation from an insulin splice-variant overexpressed in diabetes, obesity, and insulin resistance. Mol Endocrinol 19: 794–803. [crossref]
- Swanson CM, Sherer NM, Malim MH (2010) SRp40 and SRp55 Promote the Translation of Unspliced Human Immunodeficiency Virus Type 1 RNA. J Virol 84: 6748–6759.
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, et al (2005) Three-Dimensional Maps of All Chromosomes in Human Male Fibroblast Nuclei and Prometaphase Rosettes. PLoS Biology 3: 157
- Gal-Mark N, Schwartz S, Ast G (2008) Alternative splicing of Alu exons-two arms are better than one. Nucl Acids Res 36: 2012–2023.
- Meili D, Kralovicova J, Zagalak J, Bonafé L, Fiori L, et al. (2009) Disease-causing mutations improving the branch site and polypyrimidine tract: Pseudoexon activation of LINE-2 and antisense Alu lacking the poly(T)-tail. Human Mutation 30: 823–831.
- Lee J, Han K, Meyer TJ, Kim H-S, Batzer MA (2008) Chromosomal Inversions between Human and Chimpanzee Lineages Caused by Retrotransposons. PLos ONE 3: 4047.
- Warren S, Sherman SL (2001) In the Book “The Metabolic & Molecular Bases of Inherited Disease” Vol 1 Eighth Edition, Chapter 64, The Fragile X Syndrome, pg: 1257–1289.
- Ro-Choi, TS, Choi, YC (2007) A Modeling Study of Co-transcriptional Metabolism of hnRNP Using FMR1. Gene Mol Cells 23: 228–238.
- Ashley CT, Sutcliffe JS, Kunst CB, Leiner HA, Eickler EE, et al. (1993) Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat Genet 4: 244–251.
- Sittler A, Devys D, Weber C, Mandel JL (1996) Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of FMR1 protein isoforms. Hum Mol Genet 5: 95–102.
- Eichler EE, Richards S, Gibbs RA, Nelson DL (1993) Fine structure of the human FMR1 gene. Hum Mol Genet 2: 1147–1153.
- Lewin, B. (1994, 2008) RNS splicing and processing. Gene IX, Chapter 28, Pearson Prentice Hall, Pearson Education, Inc. pp 667–705.
- Ro-Choi TS, Choi YC (2009) Thermodynamic Analyses of the Constitutive Splicing Pathway for Ovomucoid Pre-mRNA. Mol Cells 27: 1010.
- Haerty W, Ponting CP (2015) Unexpected selection to retain high GC content and splicing enhancers within exons of multi-exonic lncRNA loci RNA. 21: 320–332.
- Maraia R, Sakulich AL, Brinkmann E, Green ED (1996) Gene encoding human Ro-associated autoantigen Y5 RNA. Nucleic Acids Res 24: 3552–3559. [crossref]
- Maraia RJ, Sasaki-Tozawa N, Driscoll CT, Green ED, Darlington GJ (1994) The human Y4 small cytoplasmic RNA gene is controlled by upstream elements and resides on chromosome 7 with all other hY scRNA genes. Nucl Acids Res 22: 3045–3052.
- Schwartz S, Gal-Mark N, Kfir N, Oren R, Kim E, et al. (2009) Alu Exonization Events Reveal Features Required for Precise Recognition of Exon by the Splicing Machinery. PLoS Computational Biology 5: e1000300
- Nagai K, Oubridge C, Kuglstatter A, Menichelli E, Isel C, et al. (2003) Structure, Function and Evolution of the Signal Recognition Particle. EMBO J 22: 3479–3485.
- Keene SE, Telesnitsky A (2012) cis-Acting determinants of 7SL RNA packaging by HIV-1. J Virol 86: 7934–7942. [crossref]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC genomics 8: 39
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, et al. (2010) The Nuclear- Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol Cell 39: 925.
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, et al (2003) MALAT- 1, a novel noncoding RNA, and thymosin ß4 predict metastasis and survival in early-stage non- small cell lung cancer. Oncogene 22: 8031–8041.
- Ren S, Liu Y, Xu W, Sun Y, Lu J, et al (2013) Long Noncoding RNA MALAT-1 is a New Potential Therapeutic Target for Castration Resistant Prostate Cancer. J Urology 190: 2278–2287.
- Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, et al (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73: 1180–1189. [crossref]
- Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, va den Berg E, et al. (2003) Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet 12: 1661–1669.
- Henssen AG, Koche R, Zhuang J, Jiang E, Reed C, et al. (2017) PGBD5 promotes site-specific oncogenic mutations in human tumors. Nat Genet 49: 1005–1014.
- Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, et al. (2014) The oestrogen receptor alpha- regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Communications/DOI: 101038/ncomms6383.
- Ke H, Zhao L, Feng X, Xu H, Zou L, et al. (2016) NEAT1 is Required for Survival of Breast Cancer Cells Through FUS and miR-548. Gene Regulation and Systems Biology 10: 11–17.
- Trowell H, Nagy A, Vissel B, Choo KHA (1993) Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: identification of a narrow domain containing two key centromeric DNA elements. Hum Mol Genet 2: 1639–1649.
- Waye JS, Willard HF (1989) Human ß satellite DNA: Genomic organization and sequence definition of a class of highly repetitive tandem DNA. Proc Natl Acad Sci 86: 6250–6254.
- Vissel B, Nagy A, Choo KH (1992) A satellite III sequence shared by human chromosome 13, 14 and 21 that is contiguous with alpha satellite DNA Cytogenet. Cell Genet 61: 81–86.
- Lin CC, Sasi R, Lee YS, Court D (1993) Isolation and identification of a novel tandemly repeated DNA sequence in the centromeric region of human chromosome 8. Chromosoma 102: 333–339.
- Kim J-H, Ebersole T, Kouprina N, Noskov VN, Ohzeki J-I, (2008) Human gamma-satellite DNA maintains open chromatin structure and protects a transgene from epigenetic silencing. Genome Research 19: 533–544.
- Jolly C1, Metz A, Govin J, Vigneron M, Turner BM, et al (2004) Stress-induced transcription of satellite III repeats. J Cell Biol 164: 25–33. [crossref]
- Bartlett JMS, Walters AD, Ballantyne SA, Going JJ, Grigor KM, et al. (1998) Is chromosome 9 loss a marker of disease recurrence in transitional cell carcinoma of the urinary bladder? Br J Cancer 77: 2193–2198.
- Reshmi-Skarja S, Huebner A, Handschug K, Finegold DN, Clark AJL, et al. (2003) Chromosomal fragility in patients with triple A syndrome. Am J Med Genet 117: 30–36.
- Arutyunyan A, Stoddart S, Yi S-J, Fei F, Lim M, et al. (2012) Expression of Cassini, a murine gamma-satellite sequence conserved in evolution, is regulated in normal and malignant hematopoietic cells. BMC Genomics 13: 418.
- Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ (2011) CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucl Acids Res 39: 8938–8951.
- Champlin DT, Frasch M, Saumweber H, Lis JT (1991) Characterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genes Dev 5: 1611–1621.
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, et al. (2009) Chromatin binding of SRp20 and ASF/FS2 and dissociation from mitotic chromatin is modulated by histone H3 serine 10 phoshorylation. Mol Cell 33: 450–461.
- Li X, Manley JL (2006) Cotranscriptional processes and their influence on genome stability. Genes Dev 20: 1838–1847.
- Ji X, Zhou Y, Pandit S, Huang J, Li H, (2013) SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153: 855–868.
- Chew SL, Liu H-X, Mayeda A, Krainer AR (1999) Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc Natl Acad Sci USA 96: 10655–10660.
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason, et al. (2012) Comprehensive analysis of mRNA Methylation Reveals Enrichment in 3’ UTRs and near Stop Codons Cell. doi:101016/jcell201205003.
- Xiao, W., Adhikari, S., Dahal, U., Chen, Y.-S., Hao, Y.J., Sun, B.-F., Sun, H.-Y., Li, A.,
Ping, X. -L., Lai, W. -Y., Wang, X., Ma, H.-L., Huang, C.-M., Yang, Y., Huang, N.,
Jiang, G.-B., Wang, H.-L., Zhou, Q., Wang, X.-J., Zhao, Y.-L., and Yang, Y.-G. (2016)
Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell, 61, 507–519. - Tenenbaum SA, Aguirre-Ghiso J (2005) Dephosphorylation shows SR proteins the way out. Mol Cell 20: 499–501. [crossref]
- Huang Y, Steitz JA (2005) SRprises along a messenger’s journey. Mol Cell 17: 613–615. [crossref]
- Ajiro M, Jia R, Yang Y, Zhu J, Zheng ZM (2016) A genome landscape of SRSF3- regulated splicing events and gene expression in human osteosarcoma U20S cells. Nucl. Acids Res 44: 1854–1870.
- Jumaa, H, Guénet, J-L and Nielsen, PJ (1997) Regulated Expression and RNA Processing of Transcripts from the SRp20 Splicing Factor Gene during the Cell Cycle. Mol Cell Biol 17: 3116–3124.
- Ugarkovic D (2005) Functional elements residing within satellite. DNAs EMBO Rep 6: 1035–1039. [crossref]
- Zeller P, Padeken J, van Schendel R, Kalck V, Tijsterman M, et al. (2016) Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat Genet 48: 1385–1395.
- Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD (2010) HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc Natl Acad Sci 107: 12481–12486.
- Rizvi TA, Lew KA, Murphy EC, Schmidt RD (1996) Role of Mason-Pfizer Monkey Virus (MPMV) Constitutive Transport Element (CTE) in the Propagation of MPMV Vectors by Genetic Complementation Using Homologous/Heterologous. env Genes Virology 224: 517–532.
- Ivics Z, Hackett PB, Plasterk RH, Izsv?k, Z (1997) Molecular Recognition of Sleeping Beauty, a Tc1-like Transposon from Fish, and its Transposition in Human Cells. Cell 91: 501–510.
- Ro-Choi TS, Reddy R, Henning D, Takano T, Taylor CW, et al. (1972) Nucleotide sequence of 45S RNA I of Novikoff hepatoma cell nuclei. J Biol Chem 247: 3205–3222.
- Chen W, Kalscheuer V, Tzschach A, Menzel C, Ullmann R, et al. (2008) Mapping translocation breakpoints by next-generation sequencing. Genome Res 18: 1142–1149.
- Waye,J.S. and Willard, H.F. (1987) Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from human chromosomes. Nucl. Acids Res. 15, 7549–7569
- Vissel, B. and Choo, K.H. (1987) Human alpha satellite DNA-consensus sequence and conserved regions. Nucl. Acids Res. 15, 6751–6752.
- Jolly C, Konecny L, Grady DL, Kutskova YA, Cotto JJ, et al. (2002) In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochomatin during stress. J Cell Biol 156: 775–781.
- Desmet F-O, Hamroun D, Lalande M (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucl Acids Res 37: 1–67.